INTRODUCTION
The accumulation of neuritic plaques and neurofibrillary tangles caused by amyloid-beta (Aβ) peptide deposition in the most affected brain regions, such as the medial temporal lobe and neocortical areas, is a hallmark of Alzheimer’s disease (AD), a serious neurodegenerative condition and the leading cause of dementia [1]. AD poses a major global health concern, with its prevalence and incidence continually increasing. About 50 million people worldwide suffer from dementia, with AD making up the majority of these cases—roughly two-thirds of all cases, according to recent estimates. In India, the number of AD patients is rapidly increasing, with projections indicating a potential tripling of cases by 2050 due to the aging population [2]. Despite growing evidence of the neuroprotective effects of flavonoids, their precise multi-target mechanisms in the context of AD remain poorly understood and inadequately explored using integrated computational approaches. Most existing studies focus on single-target interactions, which may not fully address the complex pathophysiology of AD. Therefore, this study aims to systematically investigate the multi-target therapeutic potential of 55 selected flavonoids using a combination of network pharmacology, molecular docking, and molecular dynamics (MD) simulations to identify key interactions with Alzheimer’s-associated proteins.
Despite years of research and investment, only five symptomatic medications (e.g., donepezil, memantine) are approved, offering limited benefits and notable side effects. Lecanemab, a monoclonal antibody targeting Aβ, has received FDA’s accelerated approval but still requires further validation for long-term safety and efficacy [3]. Consequently, alternative therapeutic approaches are being explored, including the application of bioactive compounds derived from plants [4].
Bioactive compounds, including flavonoids, alkaloids, terpenoids, and polyphenols, have been promising in combating AD. Flavonoids, naturally present in fruits and vegetables, have been shown to mitigate cellular oxidative damage and Aβ toxicity [5]. Alkaloids such as galantamine from daffodils inhibit acetylcholinesterase, enhancing cognitive function [6]. Terpenoids, such as ginkgolides from Ginkgo biloba, improve blood flow and reduce inflammation [7]. Polyphenols such as resveratrol present in grapes, possess antioxidant properties, and modulate amyloid precursor protein (APP) processing [8,9]. Curcumin, a polyphenolic compound from turmeric, exhibits anti-inflammatory and anti-amyloid effects, potentially halting AD progression [10]. Recent studies also support the role of plant-derived compounds in modulating oxidative stress via the Nrf2 pathway, including the hepatoprotective effect of olive leaf extract [11] and the protective actions of bovine lactoferrin against aflatoxin-induced damage [12]. Among these, flavonoids—abundant in fruits and vegetables—are notable for their antioxidant, anti-inflammatory, and neuroprotective activities. These properties make them promising multi-target candidates for AD intervention, particularly in counteracting oxidative stress and Aβ aggregation [13,14].
Figure 1 presents a comparative analysis of the pathological characteristics of the AD brain and a healthy brain. It emphasizes the deposition of tau protein tangles and beta-amyloid aggregates, along with the contribution of oxidative stress and inflammation, which collectively result in cell death and neuronal damage. It also illustrates the molecular mechanisms by which flavonoids mitigate AD pathology through antioxidant, anti-inflammatory effects, promotion of neurogenesis, and inhibition of beta-amyloid aggregation, along with their potential impact on neurotransmitter modulation [15].
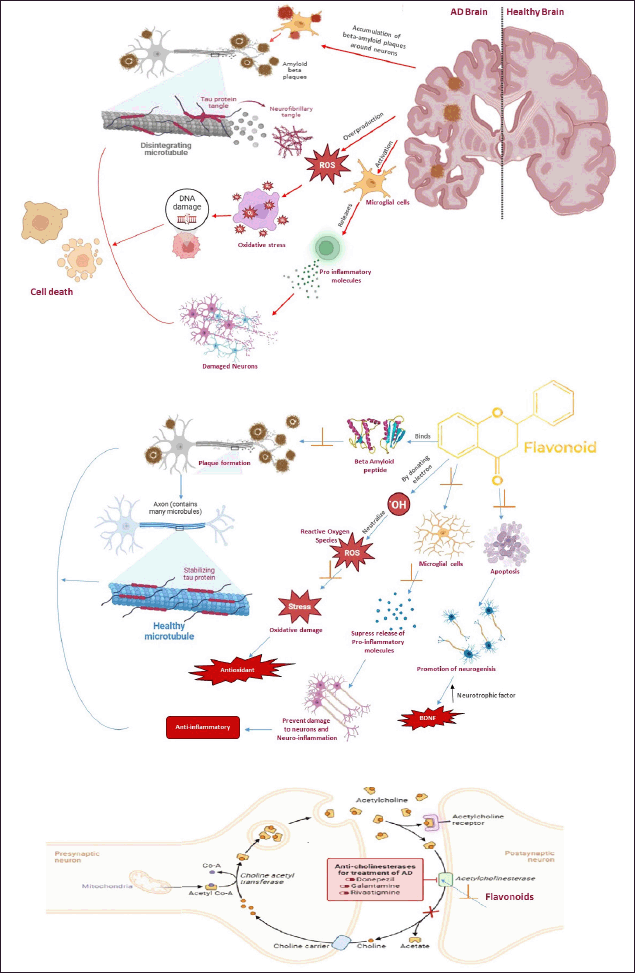 | Figure 1. Schematic representation of AD pathogenesis highlighting Aβ plaque deposition, tau hyperphosphorylation, oxidative stress, and neuronal apoptosis. The neuroprotective effects of flavonoids are illustrated through antioxidant, anti-inflammatory, and anti-amyloid mechanisms. Color coding distinguishes pathological vs. protective processes. [Click here to view] |
Beyond their role in AD, flavonoids are also acknowledged for their inflammation-modulating, anticancer, and cardioprotective effects, making them valuable in addressing multiple health conditions [16]. However, their multi-target therapeutic potential in AD remains insufficiently characterized and lacks systematic validation using integrated computational methods.
Despite substantial progress in elucidating the fundamental processes of AD pathology, existing treatment options remain limited and are predominantly symptomatic rather than disease-modifying. The plant-derived compounds, such as flavonoids, show promise since they have neuroprotective, antioxidant, and anti-inflammatory properties, yet their precise modes of action in AD remain poorly understood [17]. Traditional research often focuses on single-target therapies, which have proven insufficient in addressing the multifaceted nature of AD. Hence, there is a critical need for a comprehensive approach that explores the potential of flavonoids as multi-target agents using advanced computational tools like network pharmacology and molecular docking [18]. This research aims to close this gap by clarifying the molecular mechanisms through which flavonoids may contribute to therapeutic interventions in AD, focusing on their interactions with key AD-related targets and pathways. The selection of flavonoids in this study is strategically based on compounds that have not been extensively explored for their multi-target potential in AD, particularly in the context of integrated network pharmacology, docking, and dynamics-based approaches.
This study represents a novel integrative approach to addressing the challenges in treating AD by utilizing MD modeling, network pharmacology, and computational docking to investigate the multi-target potential of flavonoids. Unlike conventional therapeutic strategies, which often target single pathways or molecules, this research highlights the multi-targeted mechanisms of flavonoids in modulating critical AD-related targets such as β-site APP cleaving enzyme 1 (BACE1), acetylcholinesterase (AChE), glycogen synthase kinase-3 beta (GSK-3β), APP, and dual specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A). Additionally, the study uniquely identifies morin as a lead flavonoid compound with the highest binding affinity and structural stability, supported by advanced molecular simulations. This is the first study to comprehensively elucidate the association between flavonoids and DYRK1A inhibition, presenting a promising target for developing disease-modifying therapies in AD. The findings bridge computational predictions with biological relevance, paving the way for further in vitro and in vivo validation to establish flavonoids as effective multi-target agents for AD therapy.
MATERIALS AND METHODS
Network pharmacology
Targets scrutinization for flavonoids and AD related
The selection of the 55 flavonoids was based on a comprehensive review of existing literature, focusing on compounds with previously reported antioxidant and anti-inflammatory in experimental models of neurological disorders. The literature-based screening was performed using scientific databases such as PubMed, Scopus, and ScienceDirect. Particular emphasis was placed on flavonoids that have not been systematically explored against environmental neurotoxicants such as bisphenol-A and aluminum chloride, thereby aiming to address a novel research gap in the context of neurodegeneration. This strategy allowed the identification of both well-established and underexplored flavonoids with putative relevance to AD. The selected flavonoids (Apiforol, Arbutin, Aromadendrin, Astragalin, Baicalein, Butein, Calophyllolide, Cianidanol, Coutareagenin, Cyanidin, Dalbergichromene, Dalbergin, Delphinidin, Didymin, Diosmin, Equol, Eriocitrin, Eriodicytol, Fisetin, Galangin, Gallocatechin, Genistein, Genistin,Glabridin, Glycitein, Hesperetin, Hesperidin, Homoeriodicytol, Isorhamnetin, Kaempferitrin, Kaempferol, Malvidin, Morin, Myricetin, Naringenin Chalcone, Naringenin, Naringin, Narirutin, Nobiletin, Pachypodol, Pelargonidin, Peonidin, Petunidin, Phloretin, Phlorizin, Poncirin, Procyanidin, Rhamnazin, Rhoifolin, Rutin, Scutellarein, Sinensetin, Sophoraflavonoloside, Tangeretin, Wogonin) were retrieved, and their chemical structures were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/ online database, accessed on August 9, 2024).
The flavonoid targets were retrieved from SwissTargetPrediction (http://www.swisstargetprediction.ch/, 2023 version, accessed on August 10, 2024), applying a probability cut-off of >0 to retain all plausible predicted interactions. This inclusive threshold was intentionally chosen to allow a broad initial mapping of the flavonoid-target space, ensuring that no potentially relevant interactions were excluded at an early stage [19]. AD-related targets were obtained from GeneCards (https://www.genecards.org/, version 5.12, accessed on August 10, 2024) [20] using a relevance score threshold of ≥15. This score reflects high-confidence associations based on integrated genomic, transcriptomic, and proteomic evidence and is consistent with practices widely adopted in network pharmacology studies. These stringent criteria helped to enhance disease specificity while minimizing the inclusion of weak or indirect associations [21].
The overlapping targets between the selected flavonoids and AD were then identified using Venny 2.0.2 (https://bioinfogp.cnb.csic.es/tools/venny/index2.0.2.html, web tool, accessed on August 11, 2024) [22], and the resulting common targets were used for subsequent protein–protein interaction (PPI) network construction and enrichment analyses.
Construction of network and identification of PPIs
PPI networks may be predicted using the online biological database STRING (https://string-db.org/, version 11.5, accessed on August 11, 2024) [23]. From the overlapped targets, we introduced the targets of interest into the string database. The 124 targets were taken for functional enrichment analysis and retrieved the 32 targets. Again, in string, the disease-drug-target network was constructed, with the network confidence score ≥0.4, organism set as “Homo sapiens” and sent to Cytoscape (version 3.10.2, accessed on august 11, 2024) [24]. The top-ranking targets were evaluated using the cyto-Hubba plugin in Cytoscape software. This assessment identified the 10 most significant Hubba nodes based on the Maximal Clique Centrality (MCC) score.
Enrichment analysis
Functional annotation analysis, encompassing Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO), was conducted via the DAVID platform (https://david.ncifcrf.gov/summary.jsp, version 2021, accessed on August 12, 2024) [25], considering p-values < 0.05 as indicative of significant findings. The KEGG pathway assessment was utilized to infer possible mechanisms of action, and the results were graphically represented using SRplot (https://www.bioinformatics.com.cn/en, web platform, accessed on August 12, 2024) [26].
Targets and key genes of AD: correlation analysis
To verify the association between the top 10 targets and major causative targets like protein tau (MAPT) and APP [27], a correlation analysis was carried out using GEPIA (http://gepia.cancer-pku.cn/, version 2.0, accessed on August 12, 2024) [28]. The key targets, along with APP and MAPT, were entered sequentially into the search terms, with all brain tissues from GTEx selected for the correlation analysis.
Drug-likeness, physicochemical and ADMET properties of selected flavonoids
The physicochemical attributes of the selected flavonoids were obtained primarily from SwissADME and DruLito (offline tool, version 2.0, accessed on August 13, 2024). Key drug-likeness parameters were calculated using DruLito, an open-source offline tool [29] following Lipinski’s rule of five. Compounds adhering to these criteria are generally expected to exhibit favorable folding, polarity, and molecular size, enhancing their potential therapeutic efficacy [30]. The ADMET characteristics of the flavonoids were analyzed using QikProp (Schrödinger suite version 2023-1, accessed on August 13, 2024), SwissADME (http://www.swissadme.ch/, online database, accessed on August 14, 2024), ProTox-II (https://tox-new.charite.de/protox_II/, online database, accessed on August 15, 2024), and pkCSM (https://biosig.lab.uq.edu.au/pkcsm/, online database, accessed on August 16, 2024) [31].
Molecular docking
Computational docking analyses were performed using selected flavonoids against key target proteins, including APP (PDB ID: 1AAP) [32], BACE1 (PDB ID: 6EQM) [33], GSK-3β (PDB ID: 1Q4L) [34], AChE (PDB ID: 4EY7) [35], and DYRK1A (PDB ID: 4AZE). The RCSB Protein Data Bank served as the source for these protein structures [36]. Processing of crystallographic protein structures was performed using Schrödinger’s Maestro software (version 2023-1, accessed on August 25, 2024), which involved defining bond orders, incorporating hydrogen atoms, defining zero-order bonds for metal ions and disulfide linkages, and removing water molecules beyond 5 Å from the active site. At physiological pH (7.0), hydrogen bonding patterns were optimized using PROPKA. Energy minimization was conducted employing the OPLS 4 force field until the root-mean-square deviation (RMSD) exceeded 0.30. Binding sites were localized using the receptor grid generation tool, centering the grids on the coordinates of co-crystallized ligands. Ligands were pre-processed using LigPrep, where low-energy conformers were generated, and ionization states were optimized to pH 7. Docking was executed using the XP-Glide module, and ligand-target interactions were analyzed in the Maestro interface [37].
Molecular dynamics
Simulations were executed using the Desmond software suite (Schrödinger version 2023-1, simulation executed on September 20, 2024) with the OPLS 2005 force field for 150 ns to investigate structural and conformational changes in the protein–ligand complex. Initial docking outputs served as input structures, which were further refined using the System Builder tool. The complex was solvated in an orthorhombic box with simple point charge water molecules, and to maintain system neutrality, Na+ and Cl– ions were introduced. At 1.01325 bar pressure and 300 K temperature, simulations were carried out under the Normal pressure and temperature ensemble, encompassing a total of 49,000 atoms. Structural stability and conformational dynamics were evaluated using RMSD and energy fluctuation analyses, while root mean square fluctuation (RMSF) measurements were utilized to assess fluctuations in both backbone and side-chain residues [38].
RESULTS
Screening of selected flavonoids and AD targets
From the SwissTargetPrediction database, 307 targets were obtained. A total of 1,058 AD-related targets were retrieved, of which 958 non-duplicate targets were considered. Using the Venn diagram method, 124 overlapping targets were identified (Fig. 2). The lists of flavonoid-specific, disease-specific, and common targets are provided in the Supplementary File S1.
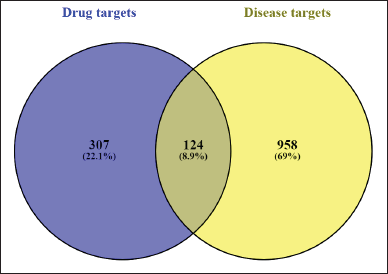 | Figure 2. Venn diagram showing the intersection of predicted flavonoid targets and AD-associated targets. Out of 307 flavonoid targets and 958 AD-related genes, 124 overlapping targets were identified using Venny 2.0.2. [Click here to view] |
Construction of the disease-flavonoid target network
The STRING database was used to analyze 124 common targets in order to create a PPI network that looked at the underlying processes of the chosen flavonoids (Fig. 3). There were 1,660 edges and 124 nodes in the built network with 612 edges expected by chance, indicating significantly more interactions than anticipated. From the functional enrichment analysis performed in Cytoscape, 32 key targets were identified (Fig. 4). We constructed a PPI network for these 32 enriched targets using the STRING database with a combined score threshold of >0.4. The constructed network was subsequently imported into Cytoscape to facilitate advanced visualization and comprehensive network analysis. To evaluate the importance of each gene within the network, the MCC ratio was calculated, using the CytoHubba and the top 10 targets—MAPT, GSK3β, APP, BACE1, CASP3, SNCA, AKT1, ACHE, GSK3A, DYRK1A, and CAPN1—were identified. The findings from the top-ranked target analysis are depicted in Figure 5, and the detailed data retrieved from functional enrichment, STRING, and the top 10 target list can be found in the Supplementary File S1.
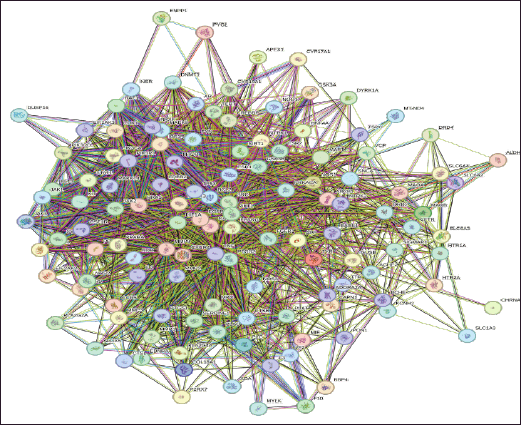 | Figure 3. STRING-derived PPI network of 124 overlapping targets. Nodes represent proteins; edges represent predicted functional associations. The interaction confidence threshold was set at ≥0.4. [Click here to view] |
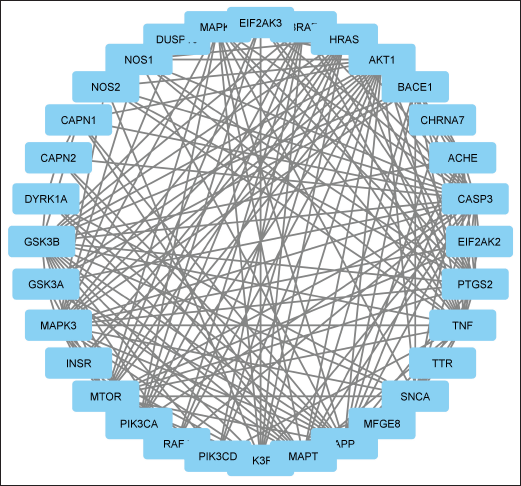 | Figure 4. PPI network of 32 targets constructed using STRING (version 11.5) and visualized in Cytoscape (version 3.10.2). Nodes represent target proteins; edges represent predicted interactions (score ≥ 0.4). Hub proteins were determined by Maximal Clique Centrality. [Click here to view] |
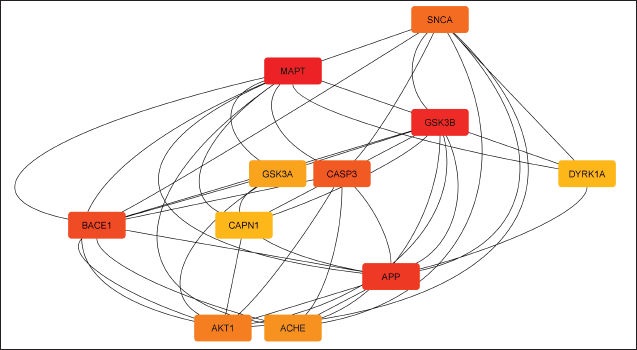 | Figure 5. Top 10 hub proteins identified via MCC analysis of the PPI network. Node color intensity reflects network importance. [Click here to view] |
GO and KEGG enrichment analysis
The DAVID database was used for the GO enrichment assessment, which classified the results into three categories: biological process (BP), cellular component (CC), and molecular function (MF). The study used the greatest incidence counts and a p-value criterion of less than 0.05 to select findings. A total of 322 GO terms were retrieved, comprising 234 related to BP, 38 associated with CC, and 50 linked to MF. From these, 30 significant GO terms were selected, with 10 each for BP, CC, and MF. The results were displayed in a bar chart using SRplot (Fig. 6). The results indicate that the potential therapeutic impact of the selected flavonoids in AD may be connected to biological mechanisms such as the G-protein-coupled receptor signaling pathway, RNA polymerase II-mediated transcription control, intracellular signal transduction, MAPK activity enhancement, protein phosphorylation, peptidyl-serine phosphorylation, inhibition of apoptotic processes, stimulation of cell proliferation, apoptosis promotion, and MFs including protein homodimerization, identical protein interaction, Adenosine Triphosphate interaction, enzyme and heme binding, macromolecular complex interaction, calmodulin association, protein tyrosine kinase activity, and protein kinase function.
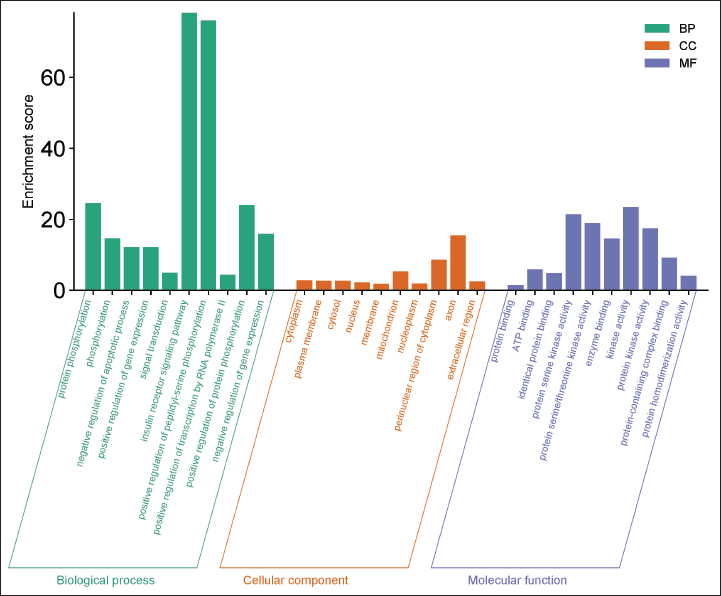 | Figure 6. GO analysis results for BP, CC, and MF categories. The top 10 terms from each category (based on p-value < 0.05) are displayed as bar graphs using SRplot. [Click here to view] |
Analysis of KEGG pathway enrichment detected 146 signaling pathways, with the top 20 ranked according to target count. According to the analysis, the targets are primarily enriched in AD-related pathways, neurodegeneration across multiple disorders, and cancer-related pathways (Fig. 7a and b). The GO and KEGG pathway data were given in the Supplementary File S1.
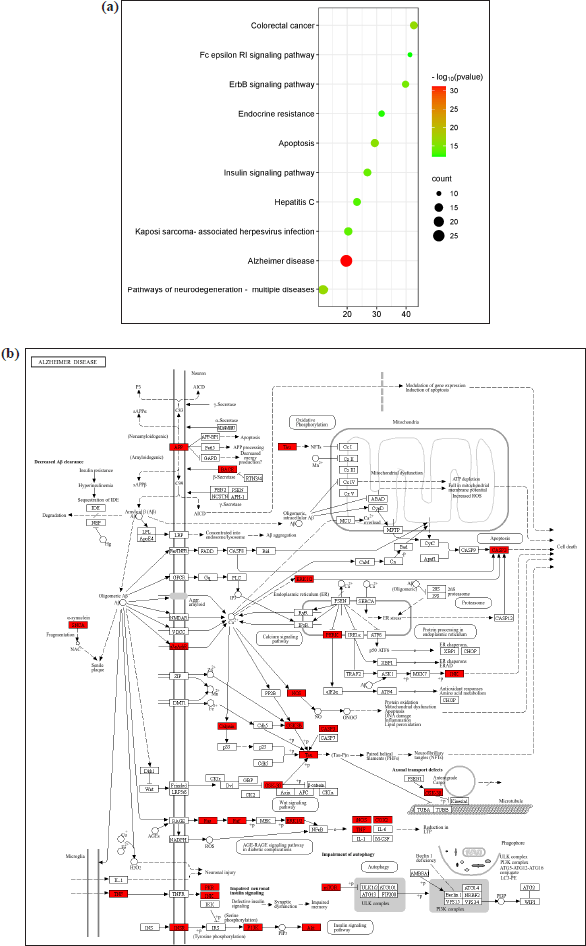 | Figure 7. (a) Bubble graph representing the top 20 enriched KEGG pathways relevant to AD, including neurodegeneration, apoptosis, and insulin signaling. Circle size indicates gene count; color reflects p-value. (b) Illustrative mapping of selected flavonoids onto AD pathway components, including interactions with BACE1, AChE, GSK-3β, APP, and DYRK1A. [Click here to view] |
Core target analysis via GEPIA
A correlation assessment was performed using GEPIA to validate the association between primary pathogenic genes, including APP and MAPT, and key target proteins. The results revealed that APP-associated targets with a p value < 0.5 for all key targets were positively correlated (Fig. 8). The causative target MPAT was also associated with key targets and with a p value < 0.5 was significantly correlated with causative targets (Fig. 9). These targets are recognized as major contributors to neurodegenerative disorders, including AD, and are implicated in worsening cognitive decline. The findings indicated that ACHE, BACE1, DYRK1A, GSK3β, and APP exhibited a stronger association with causative targets compared to MAPT, AKT1, SNCA, CAPN1, CASP3, and GSK3A. Therefore, it is hypothesized that these five targets (ACHE, BACE1, DYRK1A, GSK3β, and APP) play a crucial role in the neuroprotective and therapeutic potential of the selected flavonoids against AD.
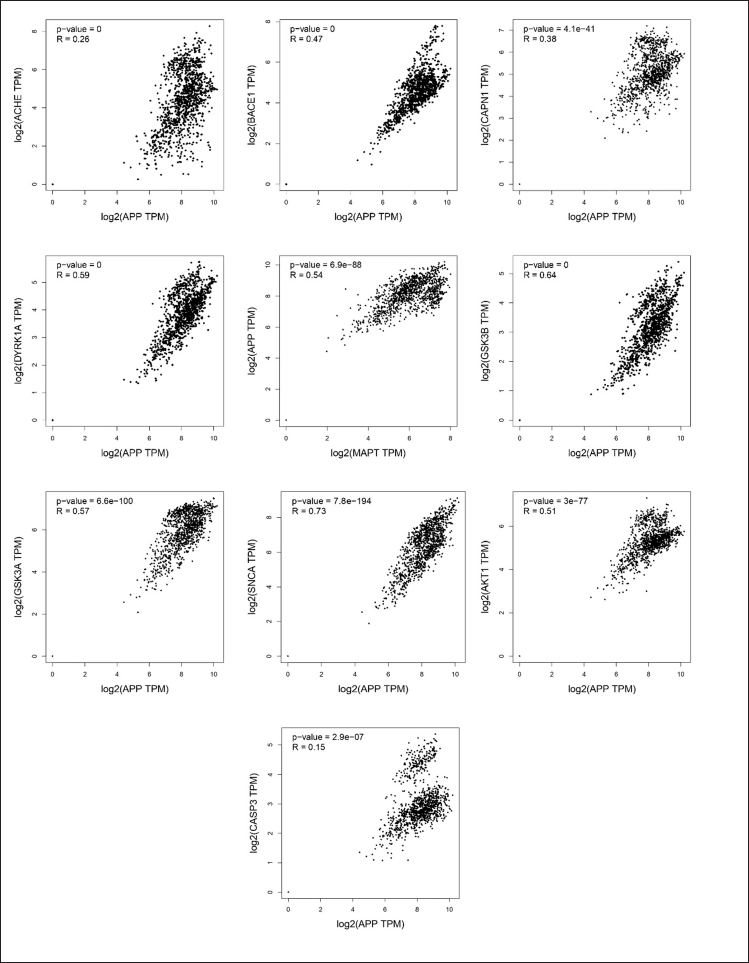 | Figure 8. Correlation plot showing expression associations between APP and key flavonoid targets in brain tissue using GEPIA (GTEx dataset). Positive correlations (p < 0.05) are emphasized. [Click here to view] |
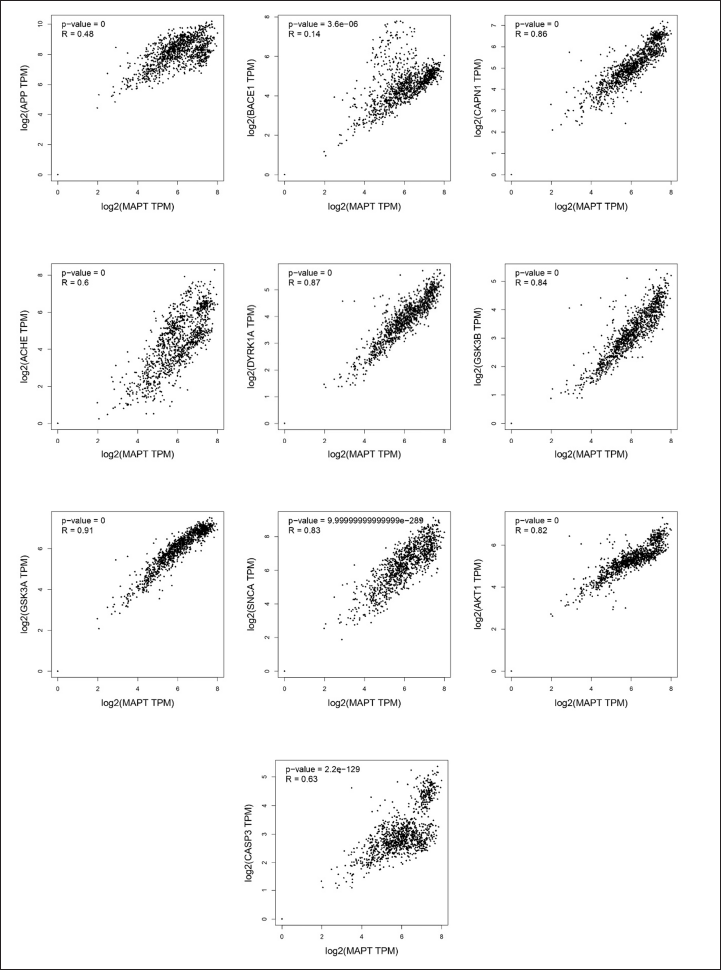 | Figure 9. Correlation plot between MAPT (tau protein gene) and the top flavonoid targets in brain tissue using GEPIA. Statistical significance (p < 0.05) indicates potential mechanistic relevance. [Click here to view] |
Physicochemical, drug-likeness, and ADMET properties of selected flavonoids
The chosen flavonoids’ physicochemical and drug-like characteristics were anticipated. Table 1 lists the important criteria such as molecular weight (MW ≤ 500 g/mol), hydrophobicity (Log P ≤ 5), topological polar surface area (TPSA ≤ 140 Å), hydrogen bond donors (HBD ≤ 5), and hydrogen bond acceptors (HBA ≤ 10). Flavonoids that did not meet Lipinski’s rule of five criteria are listed in Supplementary File S2.
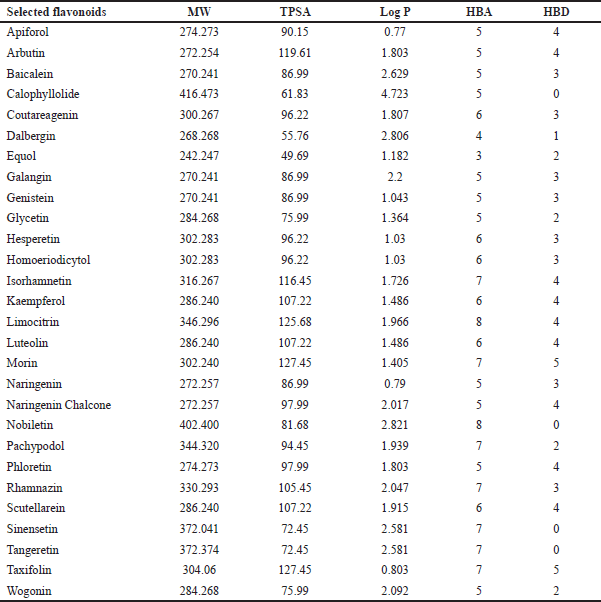 | Table 1. Physicochemical and drug-likeness properties of selected flavonoids. Parameters include molecular weight (MW), TPSA, lipophilicity (Log P), HBA, and HBD, evaluated per Lipinski’s Rule of Five. [Click here to view] |
The ADMET properties of the selected flavonoids were assessed using several key parameters, including human oral absorption (HOA), blood–brain barrier (BBB) permeability, CYP enzyme inhibition or substrate status, carcinogenicity, AMES toxicity, and LD50 in rats. None of the evaluated compounds exhibited carcinogenic properties. Flavonoids with favorable ADMET profiles are displayed in Table 2, while those with undesirable properties are listed in Supplementary File S2.
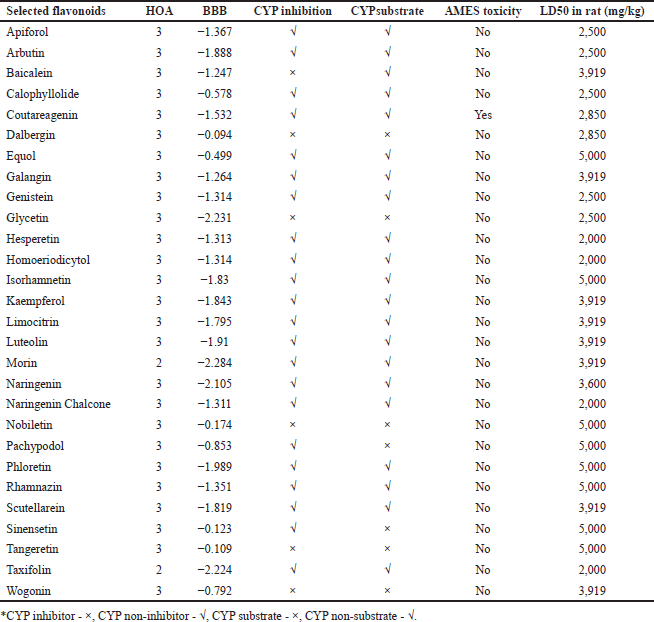 | Table 2. ADMET profile of selected flavonoids. Includes HOA, BBB permeability, CYP450 enzyme inhibition and substrate status, Ames test toxicity, and rat oral LD50. [Click here to view] |
Molecular docking
The top five selected flavonoids are shown in Table 3 against the active sites of the most enriched Alzheimer’s-associated target. Luteolin achieved the lowest XP-Glide score against AChE, suggesting strong binding affinity. Additionally, genistein recorded the lowest scores when docked with both BACE1 and APP, indicating potential inhibitory effects on these targets. Rhamnazin exhibited the lowest glide score against GSK-3β, highlighting its strong binding affinity with this protein. In comparison, Donepezil, a reference drug, had the lowest overall score against AChE but higher scores for other targets, indicating that certain flavonoids may offer comparable or even superior binding affinity to specific Alzheimer’s-associated targets. But Morin displayed the most stable interaction with DYRK1A and the strongest binding affinity among all the flavonoids (Fig. 10). The docking results of selected flavonoids and 2D and 3D images of the top five flavonoids are represented in Supplementary File S2.
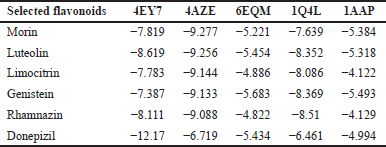 | Table 3. Docking XP scores (kcal/mol) of the top selected flavonoids in the compound-target network against the most enriched Alzheimer’s-associated target proteins. [Click here to view] |
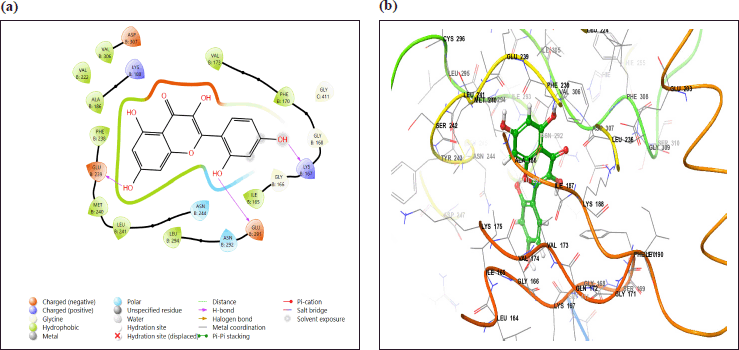 | Figure 10. (a) 3D and (b) 2D interaction diagrams of Morin with DYRK1A (PDB ID: 4AZE), highlighting key hydrogen bond interactions and hydrophobic contacts. DYRK1A is colored red, and Morin is shown in blue. [Click here to view] |
Molecular dynamics
Among all the ligands, Morin displayed the strongest binding interaction with DYRK1A in the docking studies, leading to its selection for MD simulations. The Morin-DYRK1A complex was subjected to trajectory analysis, assessing parameters such as RMSD, RMSF, and energy fluctuations. The dynamic parameters of the docked complex were carefully examined based on the plotted data. As illustrated in Figure 11, the RMSD plot confirmed the sustained stability of the docked complex throughout the 150 ns simulation, with RMSD values consistently remaining below 3 Å, validating the structural stability. Additionally, an RMSD value below 2.5 Å suggests that the protein structure remained closely aligned with the reference structure, which aligns with the findings of this study. The analysis of RMSF and energy fluctuations, as shown in Figures 12 and 13, further supports the stability and integrity of the Morin-DYRK1A complex during the simulation.
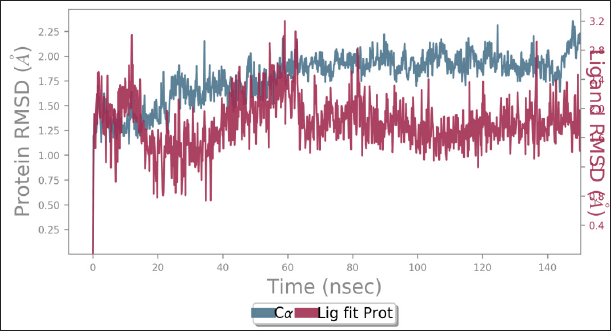 | Figure 11. Plot for RMSD of morin- DYRK1A complex during 150 ns of MD simulation. DYRK1A enzyme is shown in the red color and Morin in the blue color. [Click here to view] |
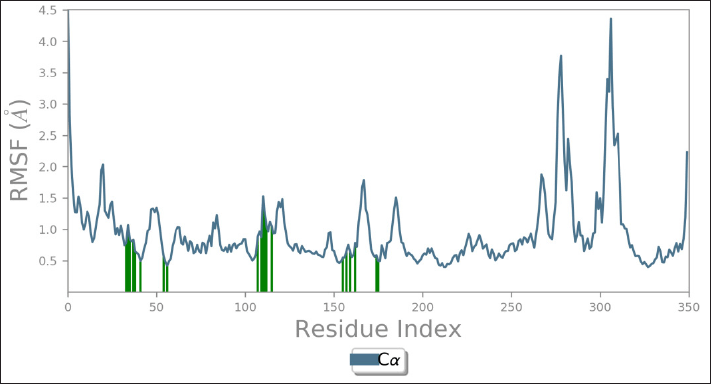 | Figure 12. RMSF analysis of the Morin–DYRK1A complex showing residue-wise flexibility, particularly at ligand-binding regions. [Click here to view] |
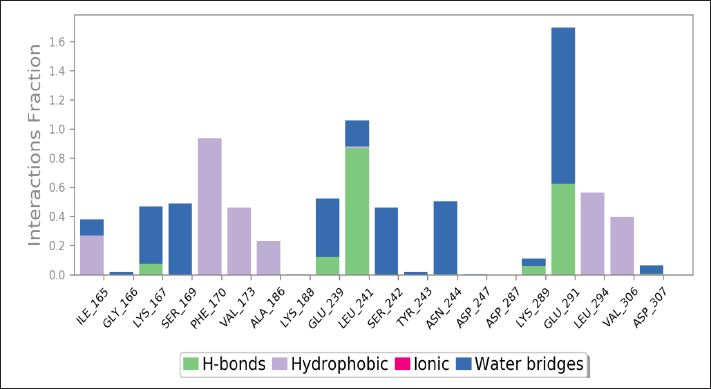 | Figure 13. Hydrogen bond analysis following a 150 ns MD simulation. Key hydrogen bonds formed by Morin with amino acid residues Leu241 and Glu291 of DYRK1A are shown. These interactions are essential for stabilizing the ligand–target complex. [Click here to view] |
DISCUSSION
In this study, we employed an integrative computational approach to evaluate the multi-target therapeutic potential of 55 flavonoids against AD. Our network pharmacology analysis identified 124 overlapping targets between the flavonoids and AD-related genes, from which 32 key proteins were prioritized through PPI analysis. Among these, five core targets—ACHE, BACE1, DYRK1A, GSK-3β, and APP—were found to play central roles in AD pathogenesis. Notably, Morin exhibited the strongest binding affinity (–9.277 kcal/mol) toward DYRK1A in docking studies, and MD simulations confirmed the stability of the Morin–DYRK1A complex (RMSD < 3 Å over 150 ns). These findings suggest that flavonoids, particularly Morin, hold promise as multi-target agents for the treatment of AD.
“AD, initially described by Alois Alzheimer in 1907, is the most common type of dementia and a major global health concern.” It is characterized by the accumulation of Aβ plaques, tau protein abnormalities, brain atrophy, and synaptic dysfunction, eventually leading to progressive cognitive decline. Despite significant research efforts, a definitive cure remains unavailable. Current treatment options, including cholinesterase inhibitors (e.g., Donepezil) and N-Methyl-D-Aspartate receptor antagonists (e.g., Memantine), provide only symptomatic relief and do not address the underlying pathology. Moreover, over 200 drugs have reached phase II trials, yet many have failed due to limited efficacy and significant side effects, underscoring the urgent need for innovative, disease-modifying therapies [39,40].
These plant-derived compounds exhibited multi-target, multi-component, and multi-pathway mechanisms, addressing the complexity of AD pathology. From 124 overlapping targets identified between flavonoids and AD-related pathways (Fig. 2), 32 key targets were refined through PPI analysis (Fig. 4). Among these, BACE1, DYRK1A, AChE, APP, and GSK3β emerged as critical targets, demonstrating strong correlations with AD markers. These results highlight the therapeutic relevance of modulating these proteins to develop effective multi-target interventions, which offer an advantage over traditional single-target therapies by simultaneously addressing multiple pathological processes of AD such as amyloid aggregation, tau hyperphosphorylation, and oxidative stress.
Flavonoids demonstrated promising binding affinities against key AD targets. Morin exhibited the strongest binding affinity for DYRK1A (–9.277 kcal/mol), followed by Luteolin (–9.256 kcal/mol) (Table 3). Notably, Donepezil, a standard AD treatment, showed superior binding to AChE (–12.17 kcal/mol) but was less effective against other targets such as BACE1 and GSK3β. These findings are consistent with research highlighting the constraints of single-target therapies in tackling AD’s multifaceted pathology [41]. For instance, BACE1, a key enzyme in Aβ production, has been implicated in neurodegeneration, as shown in transgenic models of AD [42]. DYRK1A, another critical target, regulates tau hyperphosphorylation, which exacerbates cognitive decline and neurodegeneration, making it a promising therapeutic focus [43].
Natural compounds such as resveratrol, curcumin, and ginkgolides have also shown therapeutic potential for AD. Resveratrol modulates SIRT1 activation and reduces oxidative stress, thereby improving synaptic plasticity [44]. Curcumin inhibits Aβ aggregation and exhibits anti-inflammatory properties, although its low bioavailability limits its therapeutic application [45]. Ginkgolides from G. biloba modulate inflammatory cytokines and enhance neurovascular health by improving blood flow [46]. Compared to these compounds, flavonoids such as Morin and Luteolin offer broader therapeutic potential by simultaneously targeting multiple pathways, including Aβ production, tau hyperphosphorylation, and oxidative stress.
Molecular docking and MD simulations validated the stability of flavonoid–protein interactions. The Morin-DYRK1A complex demonstrated consistent values of the RMSD less than 3 Å (Fig. 11) and variations in the minimal RMSF residue (Fig. 12), indicating robust and stable binding. Hydrogen bonding with residues such as Lys167 and Glu239 supported Morin’s potential as a therapeutic candidate. These findings highlighted DYRK1A antagonists’ role in mitigating tau hyperphosphorylation and cognitive impairments in vivo [47].
KEGG pathway and GO analysis offered further understanding of the molecular actions of flavonoids. GO annotation highlighted their role in BPs such as protein phosphorylation, apoptotic regulation, and transcriptional modulation (Fig. 6), while KEGG pathway analysis indicated significant enrichment in pathways related to neurodegeneration, insulin signaling, and programmed cell death (apoptosis) (Fig. 7). These findings support previous studies emphasizing the connection between insulin resistance and AD, particularly in the context of type II diabetes mellitus [48]. This systems-level modulation offers a mechanistic advantage over traditional single-target approaches by allowing simultaneous interference with amyloid processing (via BACE1), tau phosphorylation (via DYRK1A and MAPK), and apoptotic signaling—all of which are central to AD pathology.
ADMET profiling using QikProp, SwissADME, pkCSM, and ProTox-II demonstrated favorable pharmacokinetics and safety profiles for most flavonoids (Table 2). While Morin and Taxifolin displayed slightly lower oral absorption due to higher TPSA values, their overall ADMET profiles, including acceptable BBB permeability and non-carcinogenicity, underscore their therapeutic potential. These findings align with reports indicating that natural compounds often outperform synthetic drugs in safety and metabolic stability, making them suitable for long-term use in chronic diseases like AD [49].
Although flavonoids demonstrate significant therapeutic potential, their low bioavailability remains a critical limitation due to poor solubility, rapid metabolism, and limited BBB penetration [50]. To overcome this, several advanced drug delivery systems have been explored to enhance the pharmacokinetic and therapeutic efficacy of flavonoids. Nanoparticle-based formulations—including solid lipid nanoparticles (SLNs), polymeric nanoparticles such as PLGA and nanoemulsions—have been shown to improve solubility, protect bioactive compounds from degradation, and provide sustained release [51]. Additionally, liposomes and phytosomes have been employed to enhance cellular uptake and facilitate BBB permeability, thereby offering improved brain delivery [52,53]. For instance, curcumin-loaded SLNs have demonstrated enhanced neuroprotective effects and increased brain accumulation in AD models [54]. Furthermore, surface modifications such as PEGylation and ligand conjugation with transferrin or lactoferrin have been reported to improve BBB targeting and reduce systemic clearance, thus enhancing therapeutic efficacy [55]. These delivery platforms not only optimize the pharmacokinetic behavior of flavonoids but also provide opportunities for brain-targeted therapy in AD. Notably, several of these delivery strategies—such as curcumin-loaded SLNs and PLGA-based nanoparticles—have shown promise in preclinical models of AD, further supporting their translational relevance. Applying such approaches to flavonoids such as Morin could significantly enhance their clinical viability. Future studies should prioritize the development of morin-loaded nanocarriers and assess their therapeutic potential through in vivo validation in AD models.
This study represents a novel integrative approach to addressing the difficulties in treating AD by utilizing MD modelling, network pharmacology, and computational docking to investigate the multi-target potential of flavonoids. Unlike conventional therapeutic strategies, which often target single pathways or molecules, this research highlights the multi-targeted mechanisms of flavonoids in modulating critical AD-related targets such as BACE1, AChE, GSK-3β, APP, and DYRK1A. Additionally, the study uniquely identifies Morin as a lead flavonoid compound with the highest binding affinity and structural stability, supported by advanced molecular simulations. This interaction is particularly novel, as Morin has not been previously characterized for DYRK1A inhibition in the context of AD, and its potential role in modulating the MAPK–DYRK1A axis positions it as a unique multi-target therapeutic candidate. This is the first study to comprehensively elucidate the association between flavonoids and DYRK1A inhibition, presenting a promising target for developing disease-modifying therapies in AD. The findings bridge computational predictions with biological relevance, paving the way for further in vitro and in vivo validation to establish flavonoids as effective multi-target agents for AD therapy. While MD simulations were focused on the Morin–DYRK1A complex due to its strong binding affinity and favorable ADMET profile, this selective approach was adopted to ensure a detailed and resource-efficient investigation. Compared to Donepezil, which specifically inhibits AChE to provide symptomatic relief, Morin exhibited a broader pharmacological profile by targeting not only AChE but also key enzymes such as DYRK1A, BACE1, GSK-3β, and APP. This multi-target approach suggests that Morin could modulate various pathological mechanisms of AD, including amyloid processing, tau phosphorylation, and oxidative stress, thereby offering potential disease-modifying effects rather than symptom management alone. DYRK1A plays a critical role in AD by phosphorylating tau protein at multiple sites, contributing to neurofibrillary tangle formation and cognitive decline. The stable binding of Morin to DYRK1A indicates therapeutic relevance in targeting tau pathology, which is increasingly recognized in clinical research as a viable intervention point beyond amyloid-based strategies. However, it is acknowledged that simulating additional flavonoid-target complexes, such as Luteolin–DYRK1A, could further strengthen the mechanistic insights. Future studies will aim to incorporate these broader simulations to enhance the translational relevance of the findings. While our study focused on the identification of underexplored flavonoids, we acknowledge that comparing Morin’s binding profile with known DYRK1A inhibitors such as harmine would provide further context. This comparison was not included in the present scope but represents a valuable direction for future validation of Morin’s inhibitory potential. Similarly, we did not perform a separate MD simulation of the apo-form of DYRK1A, as the simulation focus was placed on evaluating the stability of the Morin-bound complex. Future studies may incorporate such analyses to better understand ligand-induced conformational dynamics. Future research may emphasize experimental and preclinical validation, investigate advanced delivery strategies, and explore combination therapies to fully harness flavonoids’ promise as a therapy for AD.
LIMITATION
This study was designed as an in silico exploration of the multi-target potential of flavonoids in AD. As such, no wet-lab experiments were conducted. We acknowledge that the predictive results require biological confirmation. To address this, we plan to carry out in vitro and in vivo experimental studies to validate the key findings—particularly the interaction between Morin and DYRK1A, as well as its multi-target effects relevant to AD pathology. These planned efforts will help translate the computational outcomes into potential therapeutic applications.
CONCLUSION
Taken together, Morin demonstrated the highest binding affinity and structural stability toward key AD-related target proteins in our computational analyses. This study examined the therapeutic mechanisms and potential of flavonoids—particularly Morin—in modulating AD pathogenesis through a multi-target approach, using network pharmacology, ADMET evaluation, molecular docking, and MD simulations. Given the emerging role of DYRK1A in disease progression, the Morin–DYRK1A interaction warrants further investigation. While the findings offer promising insights, they remain predictive in nature. Therefore, experimental validation is essential to establish the translational relevance of these in silico results and to assess Morin’s viability as a candidate for future drug development in AD. However, the clinical application of Morin may be challenged by its moderate oral absorption (HOA = 2), which could limit systemic bioavailability. To address this, nanoformulation-based delivery strategies—such as lipid-based nanoparticles, nanoemulsions, and PLGA carriers—could be employed to enhance solubility and BBB permeability. Additionally, developing structural analogs or prodrugs, or modifying surface characteristics of nanocarriers for targeted delivery, could further improve pharmacokinetic behavior and therapeutic efficacy. These strategies offer promising routes to translate Morin’s computational potential into a viable clinical candidate.
ACKNOWLEDGMENTS
The authors are grateful to Sri Ramachandra Institute of Higher Education and Research (DU), Chennai (India), for providing necessary support for the completion of this research work.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
CONSENT TO PARTICIPATE
All authors agree to publish the article.
SUPPLEMENTARY DATA
Supplementary files include GO and KEGG enrichment data, lists of flavonoid-specific and overlapping AD targets, 2D/3D docking images of top flavonoids, extended ADMET profiles, physicochemical properties, and docking scores. These files support and expand upon the results presented in the manuscript.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors affirm that no artificial intelligence (AI) tools were employed in the writing or editing of this manuscript, and all images remain unaltered and free from AI-based modifications.
REFERENCES
1. Aisen PS, Jimenez-Maggiora GA, Rafii MS, Walter S, Raman R. Early-stage Alzheimer disease: getting trial-ready. Nat Rev Neurol. 2022;18(7):389–99. CrossRef
2. 2024 Alzheimer’s disease facts and figures. Alzheimers Dement. 2024;20(5):3708–821. CrossRef
3. Karimi Tari P, Parsons CG, Collingridge GL, Rammes G. Memantine: updating a rare success story in pro-cognitive therapeutics. Neuropharmacology 2024;244:109737. CrossRef
4. Zhang J, Zhang Y, Wang J, Xia Y, Zhang J, Chen L. Recent advances in Alzheimer’s disease: mechanisms, clinical trials and new drug development strategies. Signal Transduct Target Ther. 2024;9(1):1–35. CrossRef
5. Naik RA, Rajpoot R, Koiri RK, Bhardwaj R, Aldairi AF, Johargy AK, et al. Dietary supplementation and the role of phytochemicals against the Alzheimer’s disease: focus on polyphenolic compounds. J Prev Alzheimers Dis. 2025;12(1):100004. CrossRef
6. Atrahimovich D, Harris R, Eitan R, Cohen M, Khatib S. Galantamine quantity and alkaloid profile in the bulbs of Narcissus tazetta and daffodil cultivars (Amaryllidaceae) grown in Israel. Metabolites 2021;11(3):185. CrossRef
7. Akaberi M, Baharara H, Amiri MS, Moghadam AT, Sahebkar A, Emami SA. Ginkgo biloba: an updated review on pharmacological, ethnobotanical, and phytochemical studies. Pharmacol Res - Mod Chin Med. 2023;9:100331. CrossRef
8. Zhang LX, Li CX, Kakar MU, Khan MS, Wu PF, Amir RM, et al. Resveratrol (RV): a pharmacological review and call for further research. Biomed Pharmacother. 2021;143:112164. CrossRef
9. Yadav E, Yadav P, Khan MMU, Singh H, Verma A. Resveratrol: a potential therapeutic natural polyphenol for neurodegenerative diseases associated with mitochondrial dysfunction. Front Pharmacol. 2022;13:922232. CrossRef
10. Islam MdR, Rauf A, Akash S, Trisha SI, Nasim AH, Akter M, et al. Targeted therapies of curcumin focus on its therapeutic benefits in cancers and human health: molecular signaling pathway-based approaches and future perspectives. Biomed Pharmacother. 2024;170:116034. CrossRef
11. Buzdar J, Shah Q, Khan M, Zaheer A, Shah T, Ataya F, et al. Hepatoprotective effects of olive leaf extract against carbon tetrachloride-induced oxidative stress: in vivo and in-silico insights into the Nrf2-NFκB pathway. J Mol Histol. 2025;56:42. CrossRef
12. Chen H, Buzdar JA, Riaz R, Fouad D, Ahmed N, Shah QA, et al. Bovine lactoferrin alleviates aflatoxin B1 induced hepatic and renal injury in broilers by mediating Nrf2 signaling pathway. Poult Sci. 2024;103(12):104316. CrossRef
13. Minocha T, Birla H, Obaid AA, Rai V, Sushma P, Shivamallu C, et al. Flavonoids as promising neuroprotectants and their therapeutic potential against Alzheimer’s disease. Oxid Med Cell Longev. 2022;2022:6038996. CrossRef
14. Forni C, Facchiano F, Bartoli M, Pieretti S, Facchiano A, D’Arcangelo D, et al. Beneficial role of phytochemicals on oxidative stress and age-related diseases. BioMed Res Int. 2019;2019(1):8748253. CrossRef
15. Kamatham PT, Shukla R, Khatri DK, Vora LK. Pathogenesis, diagnostics, and therapeutics for Alzheimer’s disease: breaking the memory barrier. Ageing Res Rev. 2024;101:102481. CrossRef
16. Hasibuan PAZ, Simanjuntak Y, Hey-Hawkins E, Lubis MF, Rohani AS, Park MN, et al. Unlocking the potential of flavonoids: natural solutions in the fight against colon cancer. Biomed Pharmacother. 2024;176:116827. CrossRef
17. Hasnat H, Shompa SA, Islam MdM, Alam S, Richi FT, Emon NU, et al. Flavonoids: a treasure house of prospective pharmacological potentials. Heliyon 2024;10(6):e27533. CrossRef
18. Bordoloi S, Pathak K, Devi M, Saikia R, Das J, Kashyap VH, et al. Some promising medicinal plants used in Alzheimer’s disease: an ethnopharmacological perspective. Discov Appl Sci. 2024;6(5):215. CrossRef
19. Gao W, Yang G, Liu X, Hu K, Pan J, Wang X, et al. Network pharmacology and experimental verification to investigate the mechanism of isoliquiritigenin for the treatment of Alzheimer’s disease. Sci Rep. 2025;15(1):4379. CrossRef
20. Zhou X, Yan Z, Wang Y, Ren Q, Liu X, Fang G, et al. Based on network pharmacology and RNA sequencing techniques to explore the molecular mechanism of Huatan Jiangzhuo decoction for treating hyperlipidemia. Evid Based Complement Alternat Med. 2021;2021(1):9863714. CrossRef
21. Patil N, Dhariwal R, Mohammed A, Wei LS, Jain M. Network pharmacology-based approach to elucidate the pharmacologic mechanisms of natural compounds from Dictyostelium discoideum for Alzheimer’s disease treatment. Heliyon 2024;10(8):e28852. CrossRef
22. Rasool K, Bhatti A, Satti AM, Paracha RZ, John P. Computational insights into the inhibitory mechanism of type 2 diabetes mellitus by bioactive components of Oryza sativa L. indica (black rice). Front Pharmacol. 2024;15:1457383. CrossRef
23. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–46. CrossRef
24. Aloliqi AA. Insights into the gene expression profile of classical hodgkin lymphoma: a study towards discovery of novel therapeutic targets. Molecules 2024;29(15):3476. CrossRef
25. Fan N, Yuan S, Hai Y, Du P, Li J, Kong X, et al. Identifying the potential role of IL-1β in the molecular mechanisms of disc degeneration using gene expression profiling and bioinformatics analysis. J Orthop Surg Hong Kong. 2022;30(1):23094990211068203. CrossRef
26. Mohanty D, Padhee S, Sahoo C, Jena S, Sahoo A, Chandra Panda P, et al. Integrating network pharmacology and experimental verification to decipher the multitarget pharmacological mechanism of Cinnamomum zeylanicum essential oil in treating inflammation. Heliyon 2024;10(2):e24120. CrossRef
27. De Simone A, Tumiatti V, Andrisano V, Milelli A. Glycogen synthase kinase 3β: a new gold rush in anti-Alzheimer’s disease multitarget drug discovery? J Med Chem. 2021;64(1):26–41. CrossRef
28. Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–60. CrossRef
29. Gnanaraj C, Sekar M, Fuloria S, Swain SS, Gan SH, Chidambaram K, et al. In silico Molecular docking analysis of karanjin against Alzheimer’s and Parkinson’s diseases as a potential natural lead molecule for new drug design, development and therapy. Molecules 2022;27(9):2834. CrossRef
30. Aqil F, Munagala R, Jeyabalan J, Vadhanam MV. Bioavailability of phytochemicals and its enhancement by drug delivery systems. Cancer Lett. 2013;334(1):133–41. CrossRef
31. Kciuk M, Malinowska M, Gielecinska A, Sundaraj R, Mujwar S, Zawisza A, et al. Synthesis, computational, and anticancer in vitro investigations of aminobenzylnaphthols derived from 2-naphtol, benzaldehydes, and α-aminoacids via the betti reaction. Molecules 2023;28(20):7230. CrossRef
32. Coburger I, Hoefgen S, Than ME. The structural biology of the amyloid precursor protein APP – a complex puzzle reveals its multi-domain architecture. Biol Chem. 2014;395(5):485–98. CrossRef
33. Ugbaja SC, Sanusi ZK, Appiah-Kubi P, Lawal MM, Kumalo HM. Computational modelling of potent β-secretase (BACE1) inhibitors towards Alzheimer’s disease treatment. Biophys Chem. 2021;270:106536. CrossRef
34. Balboni B, Tripathi SK, Veronesi M, Russo D, Penna I, Giabbai B, et al. Identification of novel GSK-3β hits using competitive biophysical assays. Int J Mol Sci. 2022;23(7):3856. CrossRef
35. Islam MT, Aktaruzzaman M, Saif A, Hasan AR, Sourov MMH, Sikdar B, et al. Identification of acetylcholinesterase inhibitors from traditional medicinal plants for Alzheimer’s disease using in silico and machine learning approaches. RSC Adv. 2024;14(47):34620–36. CrossRef
36. Chen JH, Tu HJ, Lin TE, Peng ZX, Wu YW, Yen SC, et al. Discovery of dual-specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) inhibitors using an artificial intelligence model and their effects on tau and tubulin dynamics. Biomed Pharmacother. 2024;181:117688. CrossRef
37. Samanta P, Mishra SK, Pomin VH, Doerksen RJ. Docking and molecular dynamics simulations clarify binding sites for interactions of novel marine sulfated glycans with SARS-CoV-2 spike glycoprotein. Mol Basel Switz. 2023;28(17):6413. CrossRef
38. Manandhar S, Sankhe R, Priya K, Hari G, Kumar BH, Mehta CH, et al. Molecular dynamics and structure-based virtual screening and identification of natural compounds as Wnt signaling modulators: possible therapeutics for Alzheimer’s disease. Mol Divers. 2022;26(5):2793–811. CrossRef
39. Valdez-Gaxiola CA, Rosales-Leycegui F, Gaxiola-Rubio A, Moreno-Ortiz JM, Figuera LE. Early- and late-onset Alzheimer’s disease: two sides of the same coin? Diseases 2024;12(6):110. CrossRef
40. Liu E, Zhang Y, Wang JZ. Updates in Alzheimer’s disease: from basic research to diagnosis and therapies. Transl Neurodegener. 2024;13(1):45. CrossRef
41. Andrew RJ, Fernandez CG, Stanley M, Jiang H, Nguyen P, Rice RC, et al. Lack of BACE1 S-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2017;114(45):E9665–74. CrossRef
42. Azargoonjahromi A. The duality of amyloid-β: its role in normal and Alzheimer’s disease states. Mol Brain. 2024;17(1):44. CrossRef
43. Basheer N, Smolek T, Hassan I, Liu F, Iqbal K, Zilka N, et al. Does modulation of tau hyperphosphorylation represent a reasonable therapeutic strategy for Alzheimer’s disease? From preclinical studies to the clinical trials. Mol Psychiatry. 2023;28(6):2197–214. CrossRef
44. Fang C, Xu H, Yuan L, Zhu Z, Wang X, Liu Y, et al. Natural compounds for SIRT1-mediated oxidative stress and neuroinflammation in stroke: a potential therapeutic target in the future. Oxid Med Cell Longev. 2022;2022:1949718. CrossRef
45. Rodrigues B, Ventura E, Moreira P, Resende R, Bicker J, Santos AE, et al. New low-dose curcumin derivative with therapeutic potential in Alzheimer’s disease: results from an in vitro and in vivo study in mice. Neurobiol Aging. 2025;147:105–23. CrossRef
46. Li C, Liu K, Liu S, Aerqin Q, Wu X. Role of ginkgolides in the inflammatory immune response of neurological diseases: a review of current literatures. Front Syst Neurosci [Internet]. 2020;14:45. CrossRef
47. Zhu B, Parsons T, Foley C, Shaw Y, Dunckley T, Hulme C, et al. DYRK1A antagonists rescue degeneration and behavioural deficits of in vivo models based on amyloid-β, Tau and DYRK1A neurotoxicity. Sci Rep. 2022;12:15847. CrossRef
48. Chatterjee S, Mudher A. Alzheimer’s disease and type 2 diabetes: a critical assessment of the shared pathological traits. Front Neurosci. 2018;12:383. CrossRef
49. Okella H, Okello E, Mtewa AG, Ikiriza H, Kaggwa B, Aber J, et al. ADMET profiling and molecular docking of potential antimicrobial peptides previously isolated from African catfish, Clarias gariepinus. Front Mol Biosci. 2022;9:1039286. CrossRef
50. Alshweiat A, Jaber M, Abuawad A, Athamneh T, Oqal M. Recent insights into nanoformulation delivery systems of flavonoids against glioblastoma. J Drug Deliv Sci Technol. 2024;91:105271. CrossRef
51. Prasanna P, Upadhyay A. Flavonoid-based nanomedicines in Alzheimer’s disease therapeutics: promises made, a long way to go. ACS Pharmacol Transl Sci. 2021;4(1):74–95. CrossRef
52. Juhairiyah F, de Lange ECM. Understanding drug delivery to the brain using liposome-based strategies: studies that provide mechanistic insights are essential. AAPS J. 2021;23(6):114. CrossRef
53. Talebi M, Shahbazi K, Dakkali MS, Akbari M, Almasi Ghale R, Hashemi S, et al. Phytosomes: a promising nanocarrier system for enhanced bioavailability and therapeutic efficacy of herbal products. Phytomedicine Plus. 2025;5(2):100779. CrossRef
54. Salehi B, Calina D, Docea AO, Koirala N, Aryal S, Lombardo D, et al. Curcumin’s nanomedicine formulations for therapeutic application in neurological diseases. J Clin Med. 2020;9(2):430. CrossRef
55. Teixeira MI, Lopes CM, Amaral MH, Costa PC. Surface-modified lipid nanocarriers for crossing the blood-brain barrier (BBB): a current overview of active targeting in brain diseases. Colloids Surf B Biointerfaces. 2023;221:112999. CrossRef
SUPPLEMENTARY MATERIAL
The supplementary material can be accessed at the link here: [https://japsonline.com/admin/php/uploadss/4633_pdf.pdf]