INTRODUCTION
The incidence of antibiotic resistance is recognized as a significant global public health threat. In 2020, the prevalence of cephalosporin resistance in Escherichia coli reached 47%, while methicillin resistance in Staphylococcus aureus was observed at 30% [1]. The conditions highlight the necessity for novel antibiotic agents, particularly from medicinal plants, representing a promising new therapeutic compound source, such as Jatropha multifida (commonly known as the betadine plant) [2]. Jatropha multifida, widely referred to as the coral plant, coralbush, and physic nut (a designation it shares with other species in its genus), is a Jatropha indigenous to Mexico and the Caribbean. This garden plant has been introduced to Florida and several regions in South America, Africa, the Indian subcontinent, China, and Southeast Asia. Consumption leads to gastrointestinal distress as a result of mild toxicity [3]. The castor-root plant, belonging to the Euphorbiaceae family, has a historical application in treating bleeding wounds, swelling, bone fractures, and tooth decay. Numerous authors have reported using latex in treating gastrointestinal parasitic diseases in humans and animals (anthelmintic), severe wounds linked to microbial infections, and skin inflammation [4–6].
Additionally, Das et al. [3] reported the cytotoxic and antiproliferative effects of compounds extracted from the stem of J. multifida on tumor cells [3]. The research of Dah-Nouvlessounon et al. [4] identified various phenolic and flavonoid compounds, such as 2-Hydroxybenzoic acid, o-Coumaroylquinic acid, Apigenin-apiosyl-glucoside, and luteolin-galactoside. The extracts of J. multifida exhibited bactericidal effects against various pathogens, with concentration minimally bactericidal values between 22.67 mg/ml for S. aureus and Candida albicans and 47.61 mg/ml for E. coli [4].
The necessity for substantial biomass frequently constrains the identification of bioactive compounds in plants, while these compounds are typically present in minimal quantities within plant tissues [5]. The intricate structure of these compounds can complicate chemical synthesis, resulting in high costs and inefficiencies in their production. Endophytic fungi present an alternative method for producing bioactive compounds, as they are recognized for their capacity to synthesize secondary metabolites with notable bioactivities [6].
Endophytic fungi contain various metabolites that have been identified to have antimicrobial properties in laboratory studies. These compounds include steroids, alkaloids, quinones, phenolic acids, and polyketides. Previous studies [7–10] demonstrate that endophytic fungi derived from various medicinal plants can produce antibacterial and cytotoxic compounds.
Research on the bioactive compounds produced by the endophytic fungus of J. multifida L. is scarce in the scientific literature, especially compared to other medicinal plants thoroughly investigated for their bioactivities. Jatropha multifida L. is noted for its unique secondary metabolites demonstrating diverse biological activities. This unique biochemical profile suggests the potential for novel antimicrobial compounds not found in other species. This plant has been utilized in traditional medicine, especially for treating infections; this historical application may substantiate the hypothesis that its endophytic fungi also possess antimicrobial properties. This plant is readily available and proliferates extensively in West Sumatra, Indonesia. The location of growth and the environmental conditions of this plant can directly influence the diversity of its endophytic fungi.
The urgent need for new antibiotics to treat MRSA arises from the growing issue of antibiotic-resistant bacteria and the challenges associated with their management. This study sought to identify endophytic fungi in J. multifida L. plants and assess their ability to produce antimicrobial compounds effective against harmful and drug-resistant bacteria, such as S. aureus, MRSA, and E. coli.
MATERIALS AND METHODS
Identification of sample material
Jatropha multifida L. specimens were gathered in August 2023 from Durian Taruang, Kuranji District, Padang City, West Sumatra, located at 0°55’35.4” S and 100°24’25.3” E. The collected plant parts comprised roots, stems, leaves, and fruits (Fig. 1), with a total sample weight of approximately 100 g each. After a successful identification, the specimen was preserved at the Herbarium of Universitas Andalas in Padang, Indonesia. The voucher number for plant authentication was RSM002, and the letter number was 579/K-ID/ANDA/I/2023.
 | Figure 1. The pictures of J. multifida L. (A) Leaves. (B) Fruits and flowers. [Click here to view] |
Sample preparation
Surface sterilization was conducted on healthy plant samples (roots, stems, leaves, and fruits) free from microbial infection and insect damage. The samples were washed with distilled water until all external contaminants were eliminated. The plant materials were then segmented into small pieces measuring approximately 1 × 1 cm. Surface sterilization of these segments was performed by immersing them in 70% ethanol for 30 seconds to 1 minute, followed by rinsing with sterile distilled water. The final few drops of rinse water were collected as a control. All sterilization procedures were performed under aseptic conditions. The sterilized plant segments were positioned on Sabouraud dextrose agar (SDA) plates with the damaged portion in contact with the media surface. The plates were incubated at ambient temperature (20°C–25°C) for 2–14 days. Fungal colonies exhibiting distinct morphologies were isolated and purified until a pure isolate was achieved [11].
The cultivation of pure fungi isolates in rice medium and the preparation of extracts
Pure isolates of endophytic fungi cultivated on SDA were sectioned into segments measuring 1 × 1 cm. The segments were aseptically transferred to sterile Erlenmeyer flasks containing autoclaved rice medium. The cultures were maintained at 20°C–25°C for 6 weeks, facilitating the fungal mycelium’s complete colonization of the rice medium. After incubation, the fungal cultures were macerated with ethyl acetate as the extraction solvent. The extract was concentrated with a rotary evaporator at 40°C to yield a viscous crude extract [12].
Antimicrobial activity screening
The bacterial strains utilized for screening antibacterial activity consisted of S. aureus ATCC 29213, E. coli ATCC 25922, and methicillin-resistant S. aureus (MRSA), obtained from the Microbiology Laboratory at the Faculty of Medicine, Universitas Andalas. The test of antibacterial activity was conducted utilizing the agar diffusion method. Bacterial suspensions were prepared to a turbidity standard of 0.5 McFarland and uniformly spread on the surface of nutrient agar plates. Sterile paper discs were infused with a 5% solution of endophytic fungal extract in dimethyl sulfoxide (DMSO). Chloramphenicol (Oxoid) was used as the positive control at 30-µg/disc concentration. Discs containing DMSO served as negative controls, as DMSO can solubilize all compounds without affecting bacterial cell proliferation. The discs were positioned on the surface of the pre-inoculated nutrient agar medium within Petri dishes. The plates underwent incubation at 37°C for 24 hours. Clear zones surrounding the discs were quantified to ascertain the diameter of the inhibition zones, reflecting the antibacterial activity of bioactive metabolites generated by the endophytic fungi [13].
Initial phytochemical analysis of extracts from endophytic fungi
Phytochemical screening was performed on the ethyl acetate (EtOAc) extract of the endophytic fungi exhibiting the highest antibacterial activity to ascertain the presence of diverse classes of secondary metabolites. This qualitative method sought to identify the presence of alkaloids, flavonoids, steroid–terpenoids, and phenolic compounds. The alkaloid test was conducted with Dragendorff’s reagent, resulting in a positive outcome signified by the emergence of an orange–yellow precipitate, further confirmed by Mayer’s reagent, generating a white precipitate. The flavonoid test employed cyanidin reagent, producing a positive pink hue, which was confirmed by the citroborate reagent, generating fluorescent spots under UV light at 366 nm. The steroid–terpenoid assay was performed utilizing Liebermann–Burchard reagent, yielding positive results indicated by a green–blue hue for steroids and a brown coloration for terpenoids. The phenolic test was conducted using a ferric chloride reagent, yielding a green–violet coloration indicative of a positive result [14–16].
DNA extraction and polymerase chain reaction (PCR) amplification
Both macroscopic (including colony morphology, color, and margin characteristics) and microscopic analyses were performed to characterize endophytic fungi. Molecular identification was conducted utilizing PCR with primers Internal Transcribed Spacer (ITS)-1 and ITS-4. The PCR procedure commenced with a predenaturation phase at 95°C for 1 minute, succeeded by denaturation at 95°C for 10 seconds, annealing at 52°C for 15 seconds, and extension at 72°C for 15 seconds. The three stages were reiterated for 35 cycles, after which the temperature was decreased to 4°C for final stabilization. The resultant amplification products were visualized via 1% agarose gel electrophoresis. The collected base fragments were subsequently analyzed through bidirectional Sanger DNA sequencing employing capillary electrophoresis. The acquired DNA sequences were juxtaposed with sequence data in the National Center for Biotechnology Information (NCBI) database utilizing the Basic Local Alignment Search Tool program. A phylogenetic analysis of the sequences was performed, reconstructing a phylogenetic tree via the neighbor-joining method utilizing a substitution model based on p-distance, validated by 1000 bootstrap replications, using MEGA11 (Molecular Evolutionary Genetics Analysis) software [17,18].
Liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analysis of endophytic fungi extracts
The LC-MS/MS analysis was conducted using high-performance liquid chromatography coupled with mass spectrometry. The LC-MS/MS analysis utilized an ultra-performance liquid chromatography system with a C18 column (1.8 µm; 2.1 × 100 mm) and an XEVO G2-S Q-TOF mass spectrometer. The mobile phase comprised solvent A (water with 5-mM ammonium formate) and solvent B (acetonitrile with 0.05% formic acid), running under a gradient elution system at a 0.2-ml/minute flow rate for 23 minutes. A 10-mg extract sample was dissolved in 10 ml of absolute methanol. A volume of 5 µl of this solution was introduced into the column maintained at 50°C. Mass spectrometric analysis was performed utilizing electrospray ionization in positive mode, with mass spectra acquired within 50–1,200 m/z. Data analysis was conducted utilizing MassLynx software version 4.1. Compounds were identified by analyzing the intensity of ions corresponding to the mass-to-charge ratio (m/z) of the molecular ion ([M+H])+ in comparison to the PubChem, MassBank, and ChemSpider databases. The precision of compound identification was validated by the congruence of MS/MS fragment patterns, with a tolerance of under 5 parts per million (ppm).
RESULTS AND DISCUSSION
Fungal endophytes are present in many plant species and are essential for the health of their hosts. The diversity and biological activities of these fungi are mainly unexplored. Jatropha multifida, a well-known ornamental and medicinal plant species, is found in West Sumatra, Indonesia. This investigation aimed to detect endophytic fungi that produce antibacterial metabolites in J. multifida by collecting samples of its leaves and stems. Endophytic fungi found in marine sponges, mangroves, and medicinal plants in West Sumatra, Indonesia, are the subject of an ongoing research project to identify potential new drugs [7–10,12,15,17,19,20].
Fourteen fungal isolates were successfully obtained from the roots, stems, leaves, and fruits of the J. multifida plant (Fig. 1). The endophytic fungi obtained from the roots were labeled with the code JMA, consisting of four distinct isolates. The isolates obtained from the stems were designated JMB and comprised four isolates. The isolates obtained from the leaves were labeled as JMD, amounting to five isolates. Finally, the isolate derived from the fruit was designated as JMBB, indicating a singular isolate (Fig. 2). Fungal isolates were cultivated on rice-based media. After achieving maximum growth, the fungi’s secondary metabolites were extracted with EtOAc. The extracts were then concentrated under reduced pressure until a viscous consistency was attained. The ethyl acetate extract was subsequently evaluated for antimicrobial activity.
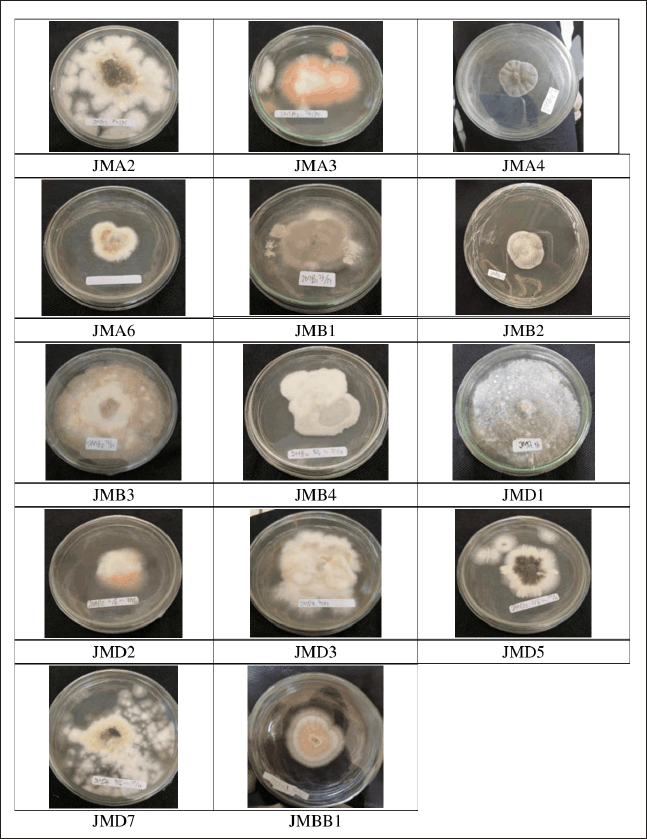 | Figure 2. The pictures of isolated fungal endophytes from J. multifida L. cultivated on SDA. [Click here to view] |
Three bacterial strains, specifically S. aureus, MRSA, and E. coli, were chosen to evaluate the antibacterial activity of fungi extracts. These strains were utilized for preliminary screening due to their availability and established testing protocols. This can be considered an initial step to identify promising fungal isolates before broadening the testing to a larger spectrum of bacteria. The selection was based on their representation of Gram-positive and Gram-negative bacteria, offering initial insight into the spectrum of activity. These strains are clinically significant and commonly associated with community-acquired and hospital-acquired infections. MRSA and E. coli are acknowledged for their multidrug resistance and are major contributors to severe infections globally.
The antibacterial efficacy of the endophytic fungi extracts was evaluated at a concentration of 5% in DMSO. Antibacterial agents on the disk diffuse into the agar, suppressing bacterial proliferation, as evidenced by a clear zone surrounding the disk. The results of the antibacterial activity screening are shown in Table 1 and Figure 3 and indicate that several extracts demonstrated differing sensitivity levels toward the test bacteria. If there is a clear zone around the disc, this suggests that the presence of secondary metabolite compounds is preventing the growth of fungi and bacteria. Seventy-nine percent of the endophytic fungi collected could inhibit the growth of S. aureus, MRSA, and E. coli strains in the experiment. This variability is due to genetic differences among fungal isolates that encode for the biosynthesis of secondary metabolites, producing various compounds with distinct antibacterial activities [21].
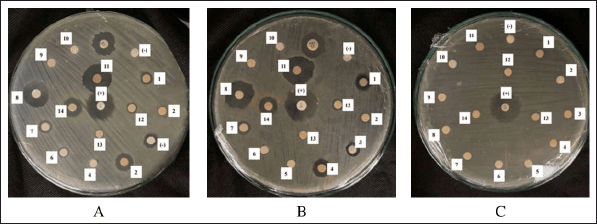 | Figure 3. The agar plate pictures of the inhibition zone of fungal isolate extract against the growth of S. aureus (A), MRSA (B), and E. coli (C). [Click here to view] |
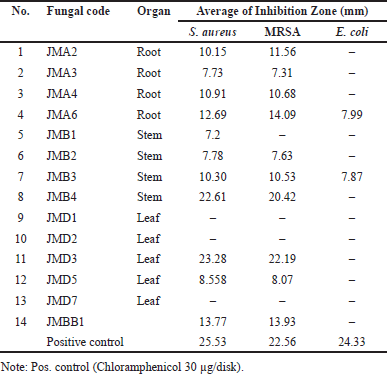 | Table 1. Antibacterial activity screening of endophytic fungi from J. multifida L. plant against pathogenic bacteria. [Click here to view] |
At a 5% concentration, seven fungal isolates exhibited significant inhibition of S. aureus and MRSA, with inhibition zones surpassing 10 mm. None of the isolates exhibited activity against E. coli (Table 2). Furthermore, the antibacterial assays indicated that the endophytic fungi isolates from J. multifida exhibit more significant potential to inhibit the growth of Gram-positive bacteria (S. aureus and MRSA) compared to Gram-negative bacteria. This phenomenon can be elucidated by the structural disparities in the cell walls of the examined bacteria, wherein Gram-positive bacteria exhibit a more simplistic cell wall composition than their Gram-negative counterparts [22]. Among all tested fungal extracts, isolates JMB4 and JMD3 demonstrated the most significant antibacterial efficacy. The JMB4 isolate exhibited an average inhibition zone of 21.79 ± 0.31 mm against S. aureus and 22.55 ± 0.71 mm against MRSA. The JMD3 isolate exhibited an average inhibition zone of 23.19 ± 0.58 mm against S. aureus and 23.37 ± 0.77 mm against MRSA (Table 2). The antibacterial efficacy of JMD3 against S. aureus and MRSA is not statistically different from that of chloramphenicol; conversely, JMB4 exhibits a significant difference from chloramphenicol for both bacteria at p < 0.05.
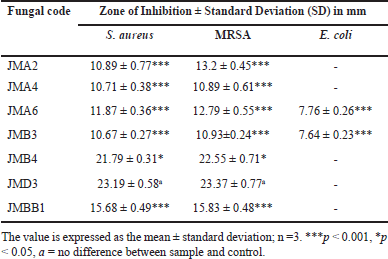 | Table 2. Antibacterial activity of selected fungal endophyte. [Click here to view] |
The extract of JMB4 and JMD3 was subsequently analyzed for their secondary metabolite composition utilizing chemical methodologies (Table 3). Prior research indicates that extracts from the stems of J. multifida L. exhibit antibacterial properties and encompass secondary metabolites categorized as alkaloids, flavonoids, steroids, phenolics, and saponins [23,24]. A comparable secondary metabolite profile was observed in the ethyl acetate extract of the endophytic fungus JMB4. Moreover, antibacterial activity has been documented in extracts from J. multifida leaves, which encompass secondary metabolites such as alkaloids, flavonoids, terpenoids, phenolics, and quinones [4,24]. The results align with the findings of the ethyl acetate extract from the endophytic fungal isolate JMD3, which demonstrated notable antibacterial activity and comprised secondary metabolites from the alkaloid, terpenoid, and steroid categories.
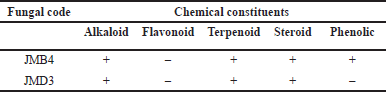 | Table 3. Phytochemical screening of EtOAc extract of GCA3 from J. multifida L. [Click here to view] |
The macroscopic examination of isolate JMB4 revealed a smooth, spherical morphology characterized by a white-cream hue and a distinctly undulating surface (Fig. 4). Conversely, isolate JMD3 exhibited clustered mycelia with a pale orange and pink hue, distinguished by wavy and tapering margins. Microscopic observations (Fig. 5) indicate that fungal isolate JMB4 possesses curved and tapered macroconidia featuring five septa. The microconidia isolate JMB4 is disk-shaped and possesses one septum. The fungal isolate JMD3 possesses curved macroconidia and oval and round microconidia resembling a false head with short monophilia. The macroscopic characteristics of both isolates were closely aligned with the descriptions of Fusarium incarnatum and Fusarium oxysporum in the literature [25].
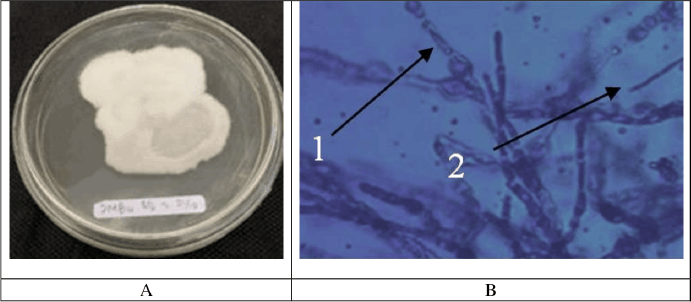 | Figure 4. Macroscopic (A) and microscopic (B) pictures of fungal isolate JMB4. [Click here to view] |
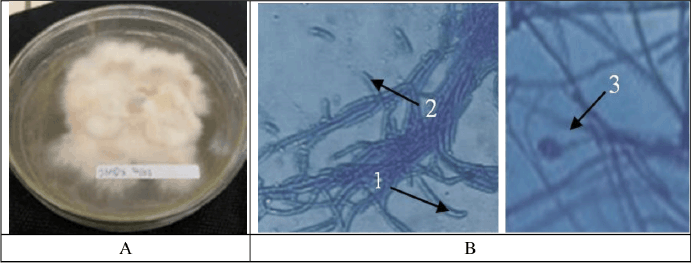 | Figure 5. Macroscopic (A) and microscopic (B) pictures of fungal isolate JMD3. [Click here to view] |
Molecular identification of fungal isolates JMB4 and JMD3 was performed utilizing PCR techniques. The DNA sequence was acquired in the ITS gene region (Fig. 6). The DNA sequences obtained were aligned with reference sequences from the NCBI database. The results were analyzed and aligned with a phylogenetic tree to validate the identification of the fungal isolates. The neighbor-joining method was utilized to reconstruct the phylogenetic tree, employing the p-distance substitution model and a bootstrap value of 1,000 replicates, analyzed with MEGA11 software (Fig. 7). Phylogenetic analysis indicates a close genetic relationship between isolate JMB4 and F. incarnatum strain JL5-2 (MT563420.1), exhibiting a percent identity of 99.81% and a query cover of 100%. Additionally, isolate JMD3 shows a close relationship with F. oxysporum isolate FoA10 (PP565979.1), with a percent identity of 99.09% and a query cover of 99% (Fig. 7).
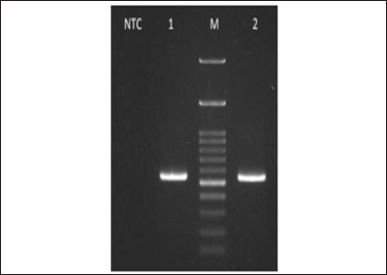 | Figure 6. Amplified isolate JMB4 (1) and isolate JMD3 (2) visualized on gel agarose 1% electrophoresis. M. 100 bp DNA ladder. NTC: Negative control amplified. [Click here to view] |
 | Figure 7. Using the neighbor-joining algorithm and 1,000 bootstraps, concatenated phylogenetic tree of isolate JMB4 and JMD3 with other Fusarium species based on internal transcribed spacer (ITS) and large subunit (LSU) rDNA region sequences. [Click here to view] |
The content of secondary metabolites in both extracts was analyzed using LC-MS/MS. A quadrupole mass analyzer detected small molecules within the mass range of 1–5,000 m/z, followed by time-of-flight analysis, which is suitable for both small and large molecules, thereby applicable to all molecular sizes. The detected masses were transformed into mass spectra, illustrating ions associated with m/z values of different compounds. The resulting MS fragments were analyzed against LC-MS/MS mass libraries from PubChem, Massbank, and prior studies.
Total ion chromatograms (TICs) for JMB4 and JMD3 extracts are shown in Figures 8 and 9. The TIC of the JMB4 extract displays several peaks, with a prominent peak observed at a retention time of 15.95 minutes. The peak exhibits a distinct adduct ion signal at m/z 994.6431 [M+H]+, correlating with the molecular formula C46H83N13O11. In analytical chemistry, specifically, mass spectrometry, adduct ions denote ions generated when a molecule interacts with another ion (typically a small ion, such as a proton, metal ion, or anion) to form a complex while preserving the fundamental structure of the molecule [26].
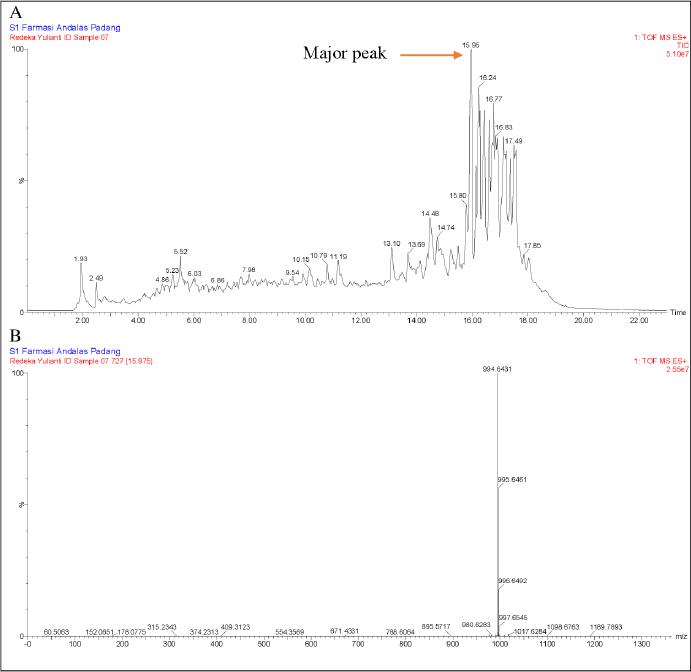 | Figure 8. LC-MS/MS data from JMB4 extract (A) and the major peak were observed at retention times of 15.95 minutes using electrospray ionization (ESI) in positive mode (B). [Click here to view] |
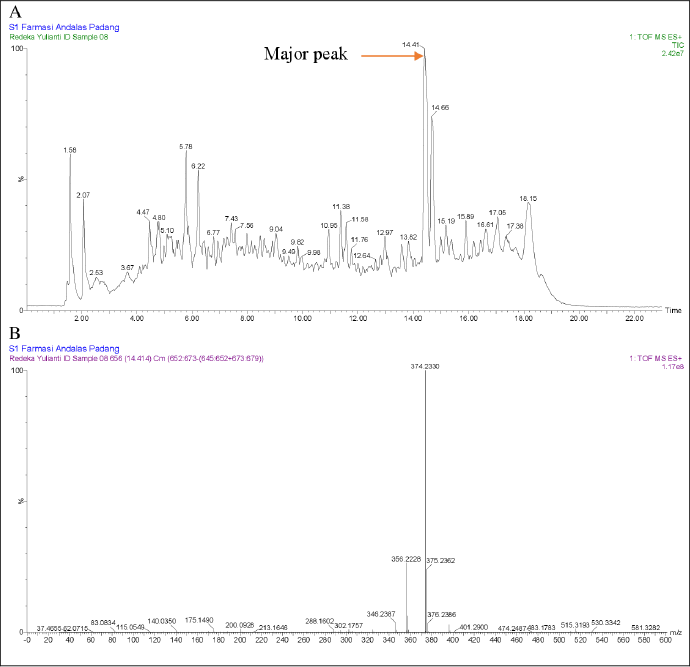 | Figure 9. LC-MS/MS data from JMD3 extract (A) and the major peak were observed at retention times of 14.41 minutes using electrospray ionization (ESI) in positive mode (B). [Click here to view] |
However, limitations in the literature precluded further identification of this compound. Conversely, the TIC of the JMD3 extract displayed multiple peaks, with the principal peak occurring at a retention time of 14.41 minutes. The peak exhibits a distinct adduct ion signal at m/z 374.2330 [M+H]+, correlating with the molecular formula C22H31NO4. The fragmentation of this ion yields several notable fragment ions, such as m/z 356.2228 [M+H]+, which aligns with the molecular formula C22H30NO3+, m/z 346.2387 [M+H]+, corresponding to C21H32NO3+, m/z 200.0928 [M+H]+, which relates to C9H14NO4+, and m/z 175.1490 [M+H]+, associated with C13H19+ (MassBank, 2021). Table 4 presents the m/z values of the parent ion and daughter ion from the major peak of the JMD3 extract, compared with the literature values for Equisetin. Based on PubChem, ChemSpider, and MassBank data comparisons, this principal peak was tentatively identified as equisetin. Initially separated from the deep-sea fungus Fusarium equiseti NRRL 5337, equisetin is an acyl tetramic acid derived from N-methylserine. Although it is not usually connected with any one strain of Fusarium, including F. equiseti, it is not widely found among a broad spectrum of fungi. The environmental conditions, the particular strain of the fungus, and the substrate on which the fungus is grown will all significantly affect the frequency of equisetin synthesis [27–29]. It demonstrates significant antibacterial activity against multidrug-resistant bacteria, particularly Gram-positive pathogens, through a novel mechanism that involves allosteric binding to biotin carboxylase [30]. Moreover, equisetin inhibits HIV-I integration and quorum sensing in P. aeruginosa. This compound was also reported to employ a distinctive dual mechanism to regulate the host-pathogen interaction in eliminating intracellular bacteria [31]. Biosynthetic researchers aim to produce equisetin because of its high activity and rarity. Research into the heritable and environmental variables that promote its growth is continuing in the hopes of improving its cultivated yield.
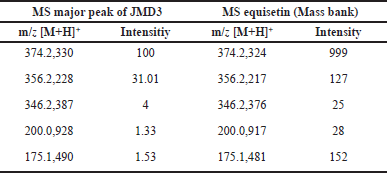 | Table 4. m/z parent ion and daughter ion from major peak JMD3 extract, which compared with the literature m/z equisetin. [Click here to view] |
Although this study has offered some preliminary insights into the antibacterial properties of equisetin from JMD3, it is essential to point out that the chemical analysis techniques utilized have significant limitations. The interpretation of the findings and the generalization of the findings are both hampered by this factor. Continuous research using enhanced isolation methods will be carried out to strengthen the findings and discover additional antibacterial compounds in JMD3 and JMB4 extracts. This will be done to address the limitations that have been identified.
CONCLUSION
Endophytic fungi from J. multifida represent a promising source for producing secondary metabolites exhibiting antibacterial properties. Fourteen endophytic fungal isolates from this plant displayed antibacterial properties against S. aureus and MRSA, with two isolates showing notable effectiveness. JMD3 demonstrates enhanced antibacterial efficacy against S. aureus and MRSA compared to JMB4. The isolate JMB4 significantly suppressed the proliferation of S. aureus and MRSA, generating inhibition zones of 21.79 ± 0.31 mm and 22.5± 0.71 mm, respectively. The isolate JMD3 exhibited inhibition zones of 23.19 ± 0.58 mm against S. aureus and 23.37 ± 0.77 mm against MRSA. Phylogenetic analysis indicated that isolate JMB4 is 100% identical to F. incarnatum, whereas isolate JMD3 exhibits a 99.09% similarity to F. oxysporum. Moreover, LC-MS/MS analysis of the JMD3 extract revealed that the predominant peak corresponded to the molecular formula C22H31NO4 and was tentatively identified as equisetin. The results of this study indicate that extracts from these fungi may provide a promising basis for creating new, effective treatments for nosocomial and skin infections, which are currently major global health issues.
ACKNOWLEDGMENT
We express our gratitude to BOPTN Universitas Andalas, Padang, Indonesia, for the financial support provided under the project titled Expertise Pathway Research (PUJK), contract number 336/UN16.19/PT.01.03/PUJK/2024.
AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS ON INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVAL
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All of the collected and analyzed data are in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Craft KM, Nguyen JM, Berg LJ, Townsend SD. Methicillin-resistant: Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. Medchemcomm. 2019;10(8):1231–41.
2. Vieira DS, de Oliveira FT, Suarez JAG, da Silva DP, Bernardo THL, de Bastos MLA. Biological activities: anti-infectious, antioxidant and healing of the vegetable species Jatropha multifida. Rev Bras Enferm. 2021;74(2):1–9.
3. Das B, Reddy KR, Ravikanth B, Raju TV, Sridhar B, Khan PU, et al. Multifidone: a novel cytotoxic lathyrane-type diterpene having an unusual six-membered A ring from Jatropha multifida. Bioorganic Med Chem Lett [Internet]. 2009;19(1):77–9. CrossRef
4. Dah-Nouvlessounon D, Chokki M, Agossou EA, Houédanou JB, Nounagnon M, Sina H, et al. Polyphenol analysis via LC-MS-ESI and potent antioxidant, anti-inflammatory, and antimicrobial activities of Jatropha multifida L. extracts used in benin pharmacopoeia. Life. 2023;13(9):1898.
5. Han T, Miao G. Strategies, achievements, and potential challenges of plant and microbial chassis in the biosynthesis of plant secondary metabolites. Molecules. 2024;29(9):2106.
6. Wang Y, Dai CC. Endophytes: a potential resource for biosynthesis, biotransformation, and biodegradation. Ann Microbiol. 2011;61(2):207–15.
7. Handayani D, Sari HC, Julianti E, Artasasta MA. Endophytic fungus isolated from Zingiber officinale Linn. var. rubrum as a source of antimicrobial compounds. J Appl Pharm Sci. 2023;13(9):115–20.
8. Handayani D, Hafiza H, Rustini R, Putra PP, Syafni N. Isolation of endophytic fungi with antimicrobial activity from medicinal plant Rhodomyrtus tomentosa (Aiton) Hassk. J Appl Pharm Sci. 2023;13(9):190–6.
9. Yulianis Y, Rustini, Supriyono A, Sandrawati N, Handayani D. Ethyl acetate extracts endophytic fungi from the medicinal tree fern cyathea contaminans (Hook) copel with antimicrobial activity. Trends Sci. 2024;21(10):1–10.
10. Handayani D, Muslim RI, Syafni N, Artasasta MA, Riga R. Endophytic fungi from medicinal plant Garcinia cowa Roxb. ex Choisy and their antibacterial activity. J Appl Pharm Sci. 2024;14(9):182–8.
11. Kjer J, Debbab A, Aly AH, Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat Protoc. 2010;5(3):479–90.
12. Handayani D, Rasyid W, Rustini, Zainudin EN, Hertiani T. Cytotoxic activity screening of fungal extracts derived from the West Sumatran marine sponge Haliclona fascia to several human cell lines: Hela, WiDr, T47D, and Vero. J Appl Pharm Sci. 2018;8(1):55–8.
13. Bauer AW, Perry DM, Kirby WMM. Single-disk antibiotic-sensitivity testing of Staphylococci: an analysis of technique and results. AMA Arch Intern Med. 1959;104(2):208–16.
14. Tiwari P, Bimlesh K, Kaur M, Kaur G, Kaur H. Phytochemical screening and extraction: a review. Int Pharm Sci. 2011;1(6):98–106.
15. Handayani D, Artasasta MA. Antibacterial and cytotoxic activities screening of symbiotic fungi extracts isolated from marine sponge Neopetrosia chaliniformis AR-01. J Appl Pharm Sci. 2017;7(5):66–9.
16. Jeyasree J, Devasena T, Sukumaran V, Women T. Phytochemical techniques—a review. World J Sci Res. 2016;1(3):67–76.
17. Sandrawati N, Hati SP, Yunita F, Putra AE, Ismed F, Tallei TE, et al. Antimicrobial and cytotoxic activities of marine sponge-derived fungal extracts isolated from Dactylospongia sp. J Appl Pharm Sci. 2020;10(4):28–33.
18. Handayani D, Artasasta MA, Safirna N, Ayuni DF, Tallei TE, Hertiani T. Fungal isolates from marine sponge Chelonaplysilla sp .: diversity, antimicrobial and cytotoxic activities. Biodiversitas. 2020;21(5):1954–60.
19. Handayani D, Artasasta MA, Mutia D, Atikah N, Tallei TE. cx Antimicrobial and cytotoxic activities screening of fungal secondary metabolites isolated from marine sponge Callyspongia sp. AACL Bioflux. 2021;14(1):249–58.
20. Handayani D, Aminah I. Antibacterial and cytotoxic activities of ethyl acetate extract of symbiotic fungi from West Sumatra marine sponge Acanthrongylophora ingens. J Appl Pharm Sci. 2017;7(2):237–40.
21. Suleiman WB. A multi-aspect analysis of two analogous Aspergillus spp. belonging to section Flavi: Aspergillus flavus and Aspergillus oryzae. BMC Microbiol. 2023;23(1):1–9.
22. Singh VK, Kumar A. Secondary metabolites from endophytic fungi: production, methods of analysis, and diverse pharmaceutical potential. Symbiosis [Internet]. 2023;90(2):111–25. CrossRef
23. Hirota BCK, Miyazaki CMS, Mercali CA, Verdan MC, Kalegari M, Gemin C, et al. C-glycosyl flavones and a comparative study of the antioxidant, hemolytic and toxic potential of Jatropha multifida leaves and bark. Int J Phytomed. 2012;4(1):1–5.
24. Rampadarath S, Puchooa D, Ranghoo-Sanmukhiya VM. Antimicrobial, phytochemical and larvicidal properties of Jatropha multifida Linn. Asian Pac J Trop Med. 2014;7(S1):S380–3.
25. Leslie JF, Summerell BA. The fusarium laboratory manual. J Chem Inf Model [Internet]. 2013;53(9):388. Available from: https://www.researchgate.net/publication/321385629_The_Fusarium_Laboratory_Manual
26. McMaster MC. GC/MS a practical user’ s guide. 2nd ed. Hoboken, NJ: John Wiley and Sons, Inc; 2008, pp. 1–6.
27. Burmeister HR, Bennett GA, Vesonder RF, Hesseltine CW. Antibiotic produced by Fusarium equiseti NRRL 5537. Antimicrob Agents Chemother. 1974;5(6):634–9.
28. Vesonder RF, Tjarks LW, Rohwedder WK, Burmeister HR, Laugal JA. Equisetin, an antibiotic from Fusarium equiseti NRRL 5537, was identified as a derivative of N-methyl-2,4-pyrollidone. J Antibiot. 1979;32(7):759–61.
29. Zhang Q, Chen S, Liu X, Lin W, Zhu K. Equisetin restores colistin sensitivity against multi-drug resistant gram-negative bacteria. Antibiotics. 2021;10(10):1263.
30. Zhang M, Wang M, Zhu X, Yu W, Gong Q. Equisetin as potential quorum sensing inhibitor of Pseudomonas aeruginosa. Biotechnol Lett [Internet]. 2018;40(5):865–70. CrossRef
31. Tian J, Chen S, Liu F, Zhu Q, Shen J, Lin W, et al. Equisetin targets intracellular Staphylococcus aureus through a host acting strategy. Mar Drugs. 2022;20(11):1–11.