INTRODUCTION
An infection with the human papillomavirus (HPV), specifically types 6 and 11, is the common cause of condyloma acuminata (CA). These viruses can lead to the development of warts or benign growths on the anal and genital regions. It is important to remember that coinfections with high-risk HPV strains can occasionally happen [1].
CA in anogenital regions and can lead to anogenital cancers, with dysplastic anal condylomas being precursors to anal squamous cell carcinoma. Immunosuppressed conditions, like due to Human immunodeficiency virus, can increase the risk of HPV infections, including CA [2]. CA poses challenges in treatment due to low efficacy and high recurrence rates. Various treatments such as cryotherapy, laser therapy, imiquimod, and surgical excision are available. However, surgical excision remains the most effective with minimal recurrence rates [3].
Piper crocatum Red betel, or Ruiz & Pav., is a species of piper that is found around the world, including Indonesia. The people of Indonesia have long been using this specific plant for a wide range of complementary and alternative medical uses. The chemical composition of red betel leaves includes flavonoids, alkaloids, tannins-polyphenols, terpenoids-steroids, and saponins. The pharmacological properties of red betel leaf, such as its anti-inflammatory, antibacterial, antifungal, antihyperglycemic, and anti-proliferative properties, have been shown in numerous research [4,5].
Molecular modeling techniques are used to bridge the gap in the understanding of the structure of bioactive chemicals at the molecular level. This technology has been used in other types of molecular systems [6–9]. The purpose of these simulations was to look into how active substances interacted with particular enzymes or receptors. For example, studies on chemicals from P. crocatum concentrated on blocking the route mechanism in CA through protein-protein inhibition. Comprehending CA requires a grasp of protein interactions and molecular simulations have shown that the p38/MAPK and PI3K/AKT signaling pathways [10] are used by mir-146a-5p to contribute to the progression of CA.
Regarding medication therapy, HPV infection is associated with multiple disorders, the most well-known being cervical cancer and CA. One of the most significant signaling pathways in the control of the growth, differentiation, and apoptosis of human cervical cancer cells is the PI3K-Akt-mTOR signaling and Apoptosis pathway. Novel approaches like high-intensity focused ultrasound and IP-10/CXCL10 gene therapy show promising results in treating CA by inducing apoptosis and inhibiting HPV [11].
Furthermore, protein interactions are studied by computer simulations, such as molecular dynamics and energy minimization, which shed light on atom movements and confirm alterations under particular circumstances [12]. These simulations support the atomic-level knowledge obtained from crystallographic investigations of known complexes and help explore protein-protein interactions, particularly in the context of weaker interactions [13]. All things considered, the combination of bioinformatics and computer simulations improves our comprehension of the molecular pathways that underlie the pathogenesis of CA.
In this work, phytochemicals from red betel leaves that may act as CA inhibitors were virtually screened. We discovered a possible protein target that may be connected to the illness and interacts promisingly with the chemical identified in red betel. As a result, these two disorders have different underlying pathways and processes.
METHODS AND MATERIALS
Retrieval of genes related to CA
By using the search term “CA” to get the GeneCards database (https://www.genecards.org), 127 candidate genes were found, of which 114 were protein coding and 13 were RNA. These genes are associated with CA. GeneCards is a comprehensive platform that provides information on gene illness connections by aggregating data from multiple public sites and academic sources. Readers can learn more about the relationships between genes and a variety of illnesses, such as Mendelian diseases, complicated diseases, and environmental diseases, thanks to this integration [14].
Protein-protein interaction network
The protein-protein interaction network was obtained by entering the list of putative genes linked to CA into the String database [15]. In biological research, String databases are frequently used, especially when looking for discovered proteins in a variety of animals and determining the possibility of protein-protein association data. These specific biological interactions include functional effects as well as direct, physical effects. Apart from the collection and reevaluation of protein-protein interactions via experimental data and the retrieval of known signaling pathways and protein complexes from other repositories, there are other ways to aid in the prediction of protein-protein interactions. These include computational transfer of interaction knowledge between organisms based on gene ontology, automated text-mining of scientific literature, systemic co-expression analysis, and identification of shared selective signals across genomes. Then, using CytoScape 3.10.0 with the String database plugin installed, we further filtered genes having a minimum interaction score of 0.999. After identifying 38 genes with an interaction score of 0.999, we constructed the interaction network.
Enrichment analysis
Using Enrichr [16], we performed enrichment analysis to find gene or protein set classes that are over-represented in the protein-protein interaction network. Enrichr includes a wide range of fundamental resources, including ontologies, pathway databases, and transcription regulation libraries. In addition, the dataset includes gene expression profiles from various cell types and tissues, as well as signatures from cells that have undergone medication treatments.
Protein and ligand preparation
The therapeutic target, X-linked inhibitor of apoptosis protein (XIAP), was selected based on the enrichment analysis. The Protein Data Bank provided the crystal structure of XIAP (rcsb.org, PDB ID: 5OQW) [17]. The protein was isolated from the co-crystallized ligand, the water molecule was removed, polar hydrogens were added, the Gasteiger charges were calculated, and chain A was recovered from the homodimer. Compounds present in red betel leaves are the ligands used in this investigation. After being sketched in MarvinSketch [18], each ligand was cleaned in 3D and stored in.pdb format.
Molecular docking
Molecular docking was implemented using AutoDock 4.2 [19]. The grid box was positioned where the co-crystalized ligand was, the exact coordinates were X = 22.345, Y = −3.445, and Z = −21.245. The grid box was set to be 40 × 40 × 40 points in size, while other parameters were set to be default. The docking itself used Lamarckian genetic algorithm and each ligand’s free binding energy was assessed. The ligand with the highest binding affinity formed a complex with XIAP and was saved in .pdb format to be analyzed further in molecular dynamics (MD) simulation.
MD simulation
Molecular docking is considered a rigid-body simulation. To analyze the physical movements of atoms and molecules in the system, we employed MD simulation using YASARA [20]. The atoms and molecules are allowed to engage in interactions over a duration of 100 ns, providing an insight into the dynamic progression of the system. The AMBER14 force field was used to estimate the forces between atoms within molecules and also between molecules. We employed the particle-mesh Ewald method to effectively manage the long-range electrostatic force. The simulation duration was 100 ns. The simulation was conducted using the constant-pressure ensemble [moles (N), pressure (P) and temperature (T)] ensemble, which maintains isothermal conditions at 298 K and isobaric conditions. A simulation cell was fabricated and subsequently filled with water, which has a density of 0.997 g/ml. To replicate physiological conditions, a concentration of 0.9% Na or Cl was introduced. The separation gap between the protein and the cell wall was established as 10 angstroms (Å). The time step utilized in the simulation was 2 femtoseconds, and snapshots of the simulation were taken at intervals of 100 picoseconds. The technique of steepest descent minimization was employed to initiate the process of structure preparation for extended MD investigations. The MD simulation was executed using a pre-established macro, md_run.mcr, which was implemented in the YASARA software. The macros md_analyse was employed to analysis the alterations in energy and residual components throughout the MD simulation. The MD simulation was employed to investigate the root-mean-square deviation (RMSD) variations, and energetic alterations.
RESULT
For the genes from Genecard with a minimum 0.999 interaction score, we used CytoScape 3.10.0 with the String database plugin added. We found 38 genes with 0.999 interaction scores and created the interaction network as Figure 1.
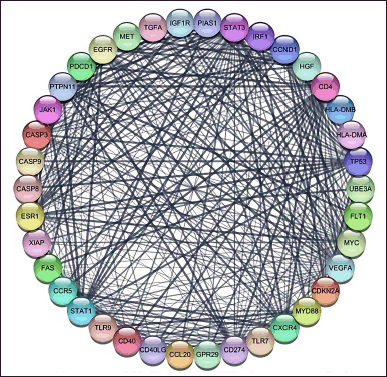 | Figure 1. CA protein-protein interaction with a confidence score of 0.99. The nodes show the proteins and the edges (line) illustrating the interactions between proteins [Click here to view] |
There are 38 genes related to the pathogenesis of CA and 96 genes that are the target of P. crocatum compounds. SwissPrediction (http://www.swisstargetprediction.ch/) [21], PharmMapper (http://www.lilab-ecust.cn/pharmmapper/) [22], GeneCards V4.12 (https://www.genecards.org/), OMIM (https://omim.org/), Pharmgkb (https://www.pharmgkb.org/), TTD (http://db.idrblab.net/ttd/), (http://www.way2drug.com/passonline/), and Pharmacology Browser 2 (PPB2) https://ppb2.gdb.tools/ are a few of the web-based servers utilized. Not all compound structures can be identified, hence many web-based platforms are used for target gene retrieval. Figure 1 displays the outcomes of the description of the number of genes involved using the webtools available at https://bioinformatics.psb.ugent.be/webtools.
Eleven genes were found to be the same between P. crocatum drugs and CA targets. Calculated numbers indicate that P. crocatum drugs target as many as 7.14% of CA genes. Thus, P. crocatum has the ability to disrupt several genes implicated in CA (Table 1). On the basis of the values of closeness centrality, degree, closeness centrality, and betweenness values, it may then be filtered for genes that typically have a close link with one another. The aforementioned grouping makes use of CytoCluster, the ClusterONE algorithm selection, and additional default-based options. After that, a number of clusters with various confidence levels will form; as seen in Figure 2, Cluster 5, which has a p-value of 0.001, is selected.
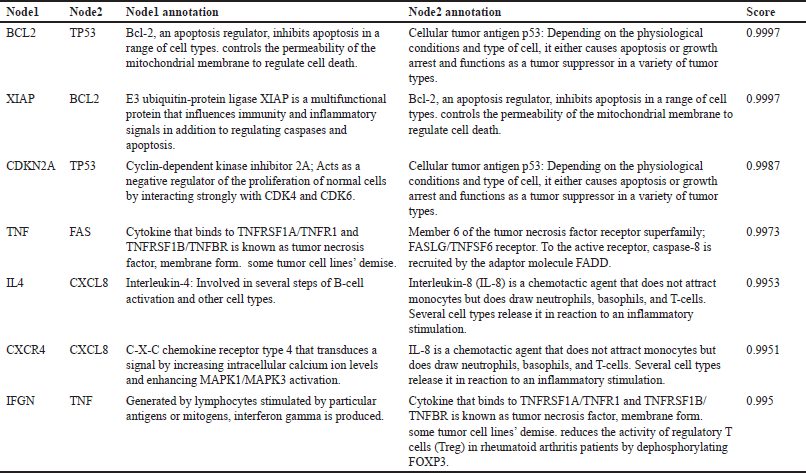 | Table 1. Genes interaction and annotation [Click here to view] |
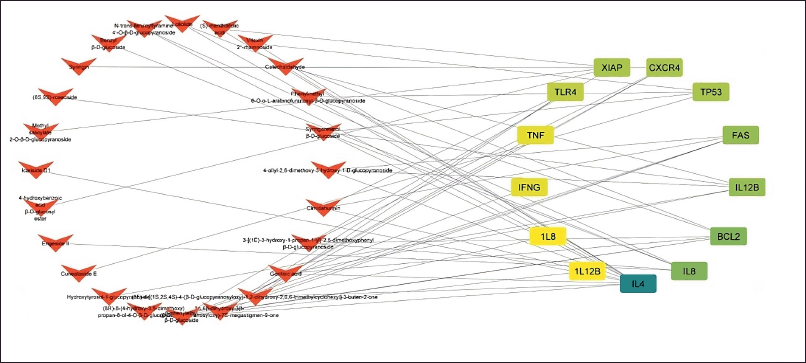 | Figure 2. Combined network construction of CA gene and active compound P. crocatum. [Click here to view] |
As illustrated in Figure 3, the gene sets that were considerably enriched were immune system response and XIAP. The biological process by cytokine production, the synthesis of interferon, the apotrophotic process, the celluler component by endosome membrane, inducing signaling, and the Clathrin-coated vesicle membrane were identified in our study’s Top 10 GO pathway enrichment analysis. The development of CA was fundamentally influenced by the C-C chemokine and Apoptototic Process signaling pathway, as well as molecular function kinase binding.
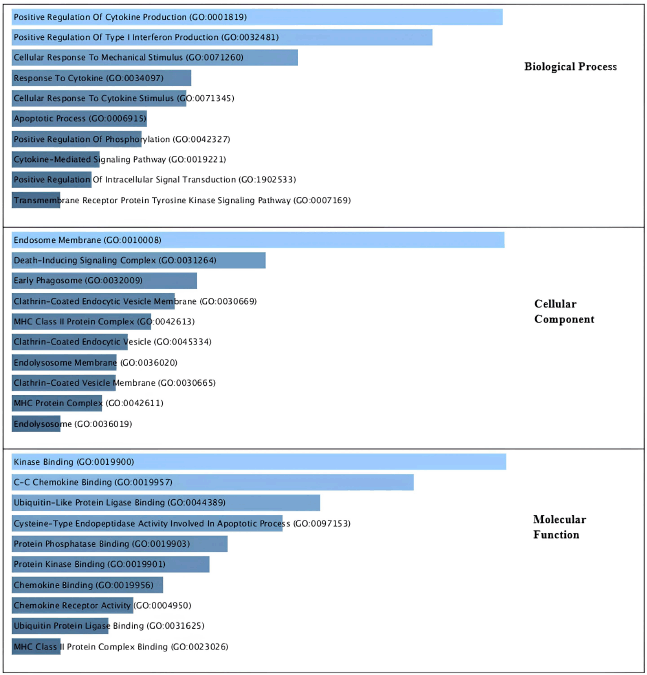 | Figure 3. Gene ontology analysis related to CA based on their biological process, cellular components and molecular function. [Click here to view] |
We used Enrichment Analysis to determine which gene classes or protein sets are overrepresented in the protein-protein interaction network, which is seen in Figure 4, based on this CA protein interaction. To learn more about the molecular relationships and biological processes connected to the CA mechanism, we examine the enriched Reactome pathways. Think about how the enriched pathways fit into the biology of the known CA mechanism, whether they provide any new information, and how they relate to the immune system, cytokine signaling, and the intrinsic apoptotic pathway.
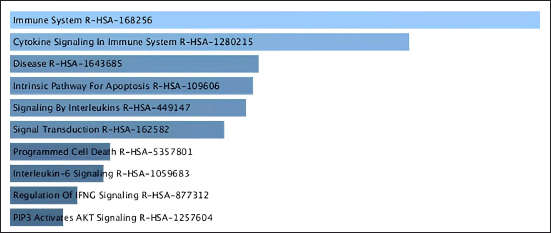 | Figure 4. Pathway analysis related to CA based on Reactome database. [Click here to view] |
By conducting enrichment analysis focusing on mechanisms and Reactome pathways, we can gain valuable insights into the biological processes underlying the CA mechanism of interest and identify potential therapeutic targets or biomarkers. Figure 4. showed that the mechanisms of CA have six signaling pathways: Immune system R-HAS, Cytokine signaling, R-HAS, Apoptosis, Interleukin, and Transduction.
The gene is then examined in terms of its enrichment and gene ontology to determine the most likely pathways and to discover that the STRING platform (https://string-db.org/), DAVID: Functional Annotation Tools (ncifcrf.gov), and metascape.org can be used to analyze interactions between genes and proteins. These resources are connected to other gene ontology servers, including the KEGG Pathways database (https://-www.genome.jp/) [23], Human References Interactome (http://www.interactome-atlas.org/), and the GAD Disease Genetic Association Disease Database (https://geneticassociationdb.nih.gov/). Using KEGG pathways, the team is able to find overrepresented genes in networks related to obesity. The apoptotic process and the PI3K signaling route are the two signaling pathways identified by KEGG. After that, every signal route is examined, and as Figure 5 below illustrates, the Apoptotic by KEGG pathway is the most likely pathway connected to CA.
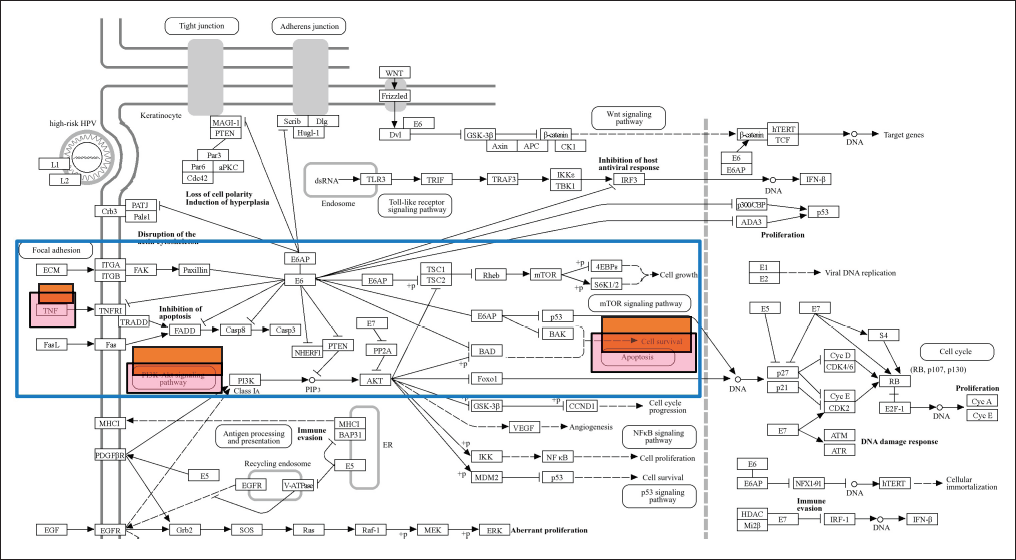 | Figure 5. Gene signaling pathway of HPV infection from KEGG database. Red box are mechanism pathway of CA with compounds of P.crocatum. [Click here to view] |
Members of the inhibitor of apoptosis (IAP) family include cellular inhibitors of apoptosis protein 1 and XIAP. Although XIAP’s precise function in CA (genital warts) is not well understood, its participation in the regulation of apoptosis and cell survival, as well as its recognized actions in other settings, allow us to speculate about its possible processes. XIAP is a strong apoptosis inhibitor. By attaching to and inhibiting caspases, especially caspase-3, -7, and -9, which are important apoptotic process executors, it carries out its anti-apoptotic activity. XIAP may help CA cells survive by averting HPV-infected cells from going through apoptosis.
Validation and molecular modeling of XIAP with compounds in P. crocatum
A grid box is a crucial component used for the validation and optimization of docking parameters. It helps in defining the search space within the protein-ligand binding site where the docking algorithm will explore potential binding poses of the ligand molecule. The grid box is typically defined based on the coordinates of the binding site of the target protein. It is a 3D box with specified dimensions that encompass the region where the ligand is expected to bind. The size and dimensions of the grid box should be carefully chosen to cover the entire binding site while minimizing unnecessary empty space. This docking of XIAP with compound in P. crocatum showed in Table 2. The grid box space is 40 × 40 × 40 with a binding energy of native ligand −12.72 and RMSD 1.64. This often requires knowledge of the binding site residues and the size of the ligand molecules being docked.
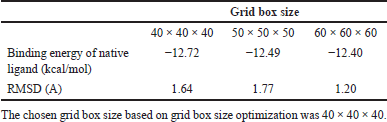 | Table 2. Grid box size optimization [Click here to view] |
Docking Xiap (XIAP) with a compound from P. crocatum involves predicting the binding mode and affinity of the compound within the binding site of the Xiap protein as Table 3.
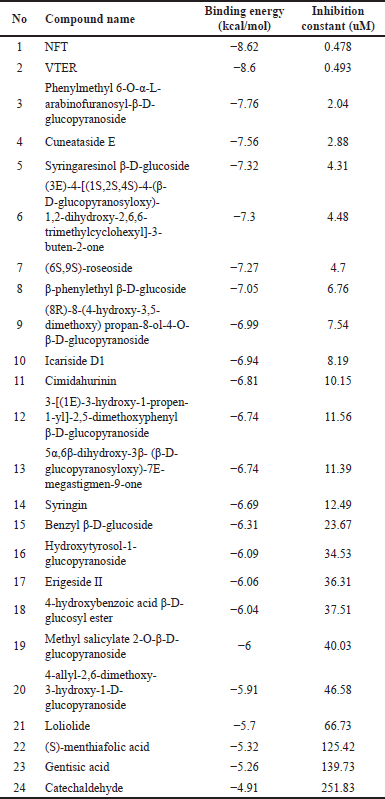 | Table 3. Docking result of XIAP with compounds in P. crocatum [24] . [Click here to view] |
Twenty-four compounds found in P. crocatum extract were subjected to molecular docking. N-trans-feruloyltyramine 4′-O-β-D-glucopyranoside (NFT) was found to have the lowest binding energy. Figure 6 showed that two compounds from P. crocatum as Candidate compounds for CA inhibitor via XIAP.
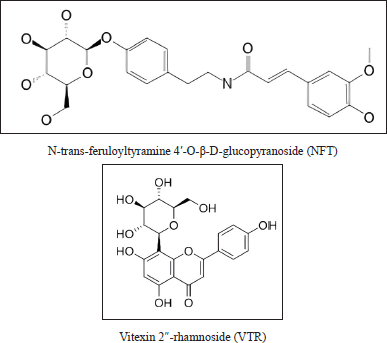 | Figure 6. Active compound in P. crocatum as XIAP inhibitor. [Click here to view] |
Retrieve the docking results generated by Autodock 6.2. This will include information on the binding poses of NFT and Vitexin 2″-rhamnoside (VTR) compounds of P. crocatum etanol extract within the binding site of XIAP, as well as associated docking scores or energies. Typically, ΔG_bind (the binding free energy) can be broken down into multiple contributing factors, such as ΔG_solvation, ΔG_entropy, ΔG_strain, and ΔG_dock. Every term adds to the total binding affinity and denotes a distinct facet of the ligand-protein interaction.NFT-XIAP and VTR-XIAP have values of −8.68 and −8.6 Kcal/mol, respectively, in Table 2.
Examine the several kinds of interactions that arise between VTR and NFT using XIAP, including electrostatic, hydrophobic, and hydrogen bonding interactions. XIAP docking studies have been thoroughly investigated in a number of research publications. Different methods, including molecular docking and MD simulations, have been used to find putative inhibitors of XIAP. The goal of these investigations is to find new chemicals that target CA [24] and XIAP, two essential proteins for cancer cell survival, to cause apoptosis.
MD simulation of XIAP-NFT
Molecular docking cannot adequately account for the structural flexibility of proteins. To further elucidate the critical interactions between small molecule medicines and the XIAP protein, MD simulation was performed using the best molecular docking model available for this inquiry.
In-depth MD simulations are run on the ligand-protein complex to investigate the conformational space and calculate the free energy change brought about by ligand binding. In MD trajectory analysis, RMSD and root mean square fluctuation (RMSF) are the most often used indices. The RMSD increases with conformational instability. Figure 7 shows how, during the transportation process, the tiny molecule varied at first and then gradually tended to be constant. In the 100 ns MD simulation, the RMSD between the four chemicals and the XIAP protein attained a relative equilibrium condition. These four compounds were found to be approaching the location where the XIAP protein is expected to be, suggesting that the small molecule was successfully integrated with the protein surface and encouraging stability between them. RMSF can be used to observe the allosteric of local areas during the simulation process. With increasing RMSF, the degree of conformational shift in the residue rises. Two was selected as the cutoff point. Figure 8 shows the wide variation in residues 250–255, 273–275, and 310–315 in the interaction between NFT and XIAP protein. Protein-ligand interactions were classified into four groups. They comprised hydrogen bonds, ions, water bridges, and hydrophobic interactions. The binding of the Delta strain complexes and the binding of NFT with XIAP of phi-phi stalking were primarily mediated by H-bonds. When XIAP was bound by the Lambda strain, hydrophobic contact was seen.
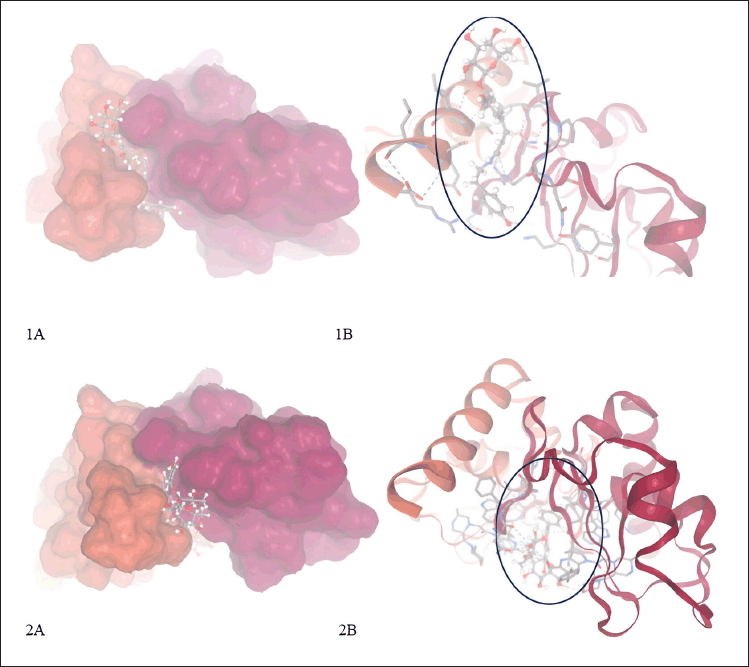 | Figure 7. Complex from molecular docking XIAP with compounds in P. crocatum. 1A. 3D and surface area and cavity of complex XIAP-NFT; 1B. 2D and binding hydrogen of XIAP-NFT; 2A. 3D and surface area and cavity of complex XIAP-VTR; 2B. 2D and binding hydrogen of XIAP-VTR. [Click here to view] |
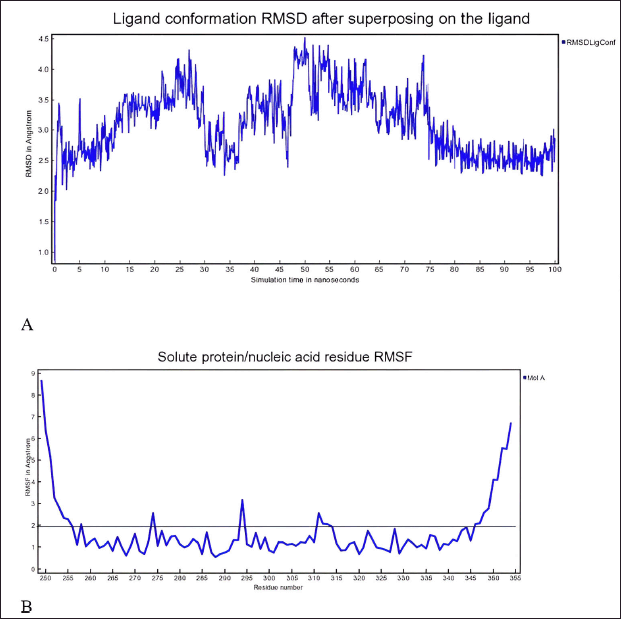 | Figure 8. A) RMSD and B) RMSF of complex XIAP and N-trans-feruloyltyramine 4′-O-β-D-glucopyranoside during 100 ns MD simulation the ligand-protein complex is subjected to extensive MD simulations to explore the conformational space and calculate the free energy change associated with ligand binding. [Click here to view] |
DISCUSSION
The primary symptom of CA is recurrent, frequent genital warts caused by HPV-infected cells. Vascular distension and papillomatosis, as described in Genomics 114 (2022) 1103035, are the hallmarks of CA pathogenesis. Nor is the precise process well understood. To investigate the marker and the impact of P. crocatum as an inhibitor of CA markers, the current study used bioinformatic techniques. 38 genes with 0.99 interaction and annotation genes as Table 1 are displayed in Figure 1. BCL2, XIAP, CDKN2A, TNF, IL4, CXCR4, IFGN, TLR4, FAS, and IL12B are the 11 hub genes that we have found.
In the event of a general viral skin infection, pattern recognition receptors that identify viral DNA or RNA [25] are expressed by keratinocytes, which are regarded as sentinels in the innate immune response. These receptors, referred to as Toll-like receptors (TLRs), are produced in endosomes or on the surface of cells and are able to detect molecular patterns linked to pathogens. Endosome-resident TLRs stimulate distinct immune pathways by upregulating the production of type I interferon (IFN) and pro-inflammatory cytokines through IFN regulatory factors and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) [26]. IFN, NFkB, and TLRs are included in the gene marker of CA, according to PPI Figure 1.
These dendritic cells exhibit surface molecules, such as CD83 and MHC (class I and II), which are necessary for antigen presentation, together with costimulatory molecules, like CD80 and CD86, and release pro-inflammatory cytokines during maturation, following virus detection. Through the CCR7 receptor, these cells become sensitive to the chemokines CCL19 and CCL21, which permits them to migrate to the lymph nodes. Following migration, immature T cells that are directed towards the infected region are activated by Langerhans cells, hence triggering an adaptive immune response [27,28]. When an intracellular pathogen is encountered, the T cell response happens in two stages: 1) strong antigen presentation that causes antigen-specific T cell growth, and 2) apoptosis-induced constriction of the activated T cells [29].
One essential process that keeps the immune system in a state of equilibrium is apoptosis. The activation of the caspase family of cysteine proteases, which can be triggered by either the cell’s intrinsic or extrinsic routes [30], is what causes apoptotic cell death. In the latter stages of the CD8 T cell response’s growth phase, caspases are activated [31]. Numerous routes can lead to apoptosis, and in activated T cells, non-apoptotic pathways can also result in cell death [32]. In a more recent study, it was demonstrated that cell autonomous caspase-8 limits CD8 T cell proliferation in response to viral infection without affecting the immune response’s contraction [33].
We used a pharmacology browser to investigate the impact of P. crocatum as a marker derived from this Hub gene. Not all compound structures can be identified, hence many web-based platforms are used for target gene retrieval. The mechanism by which P. crocatum functions as a CA inhibitor by targeting XIAP is illustrated in Figure 2. The family of endogenous caspase inhibitors and cell death regulators is known as IAPs [34]. To avoid cell death processes, cancer cells frequently express high quantities of IAPs [35,36]. The most effective inhibitor of caspase 3, 7, and 9 [37,38] among all the IAP family members is XIAP.
Figure 4 shows how XIAP and BCL2 helped the apoptotic process. that by controlling PTEN concentration and compartmentalization, XIAP functions as an E3 ubiquitin ligase for PTEN and stimulates Akt activation. CA overexpression of XIAP is linked to the progression of the illness [39]. Furthermore, research has demonstrated the involvement of other immune-related genes and pathways in CA, including the overexpression of IL7 [40] and the downregulation of GZMB, IFNG, IL12B, and IL8. Furthermore, via modifying HPV through the UBE3A and IGF-1R pathways in cervical cancer cells, microRNA-375 has been found to be a putative anti-CA mechanism [41]. Furthermore, it has been noted that individuals with CA have altered expression levels of MyD88 and TLRs, which suggests an immunological dysregulation and increased immunosuppression [42]. Lastly, it has been shown that Yes-associated protein (1) activates the EGFR pathway [43], which in turn promotes the development of keratinocytes in CA.
Researchers have identified promising natural compounds like NFT and VTR as potential lead compounds for XIAP inhibition as Table 3 from P. crocatum. These findings are based on molecular simulation and analysis of the binding modes and affinities of XIAP inhibitors with RMSD <2 A as Table 2. Asn 252, Thr254, Arg258, Gly305, Ser347, Leu348, and Pro352 of the XIAP domain were necessary for compound binding, according to the binding hydrogen analysis of bonds between this NFT-XIAP domain in the final structure of complexes (Fig. 8; 1A and B). This graphic demonstrated that the XIAP cavity site is huge and that the NFT cavity configuration is the same as the XIAP cavity. Examine the NFT and VTR’s binding positions within the binding cavity to comprehend their orientation and interactions with the XIAP protein residues. Determine the important interactions between the ligand and protein, as shown in Figure 8 1B and B, such as hydrogen bonds, hydrophobic contacts, salt bridges, and π-π stacking interactions. The interactions between XIAP and other ligands have been predicted thanks in large part to molecular docking techniques, which have yielded important information for the creation of anti-cancer treatments that specifically target XIAP. The C-glycosyl flavones that VTX produces have significant health benefits for people. Research has indicated that vitexin exhibits antitumor activity in preclinical models of ectopic development of various cancer cells, such as those found in the breast, prostate, liver, and cervical regions. It has been demonstrated that the C-linked glycosidase flavones vitexin and isovitexin block alpha-glucosidase, the enzyme that breaks down carbohydrates into sugar.
The family of apoptosis proteins includes the XIAP protein. Comprising three Baculoviral IAP Repeat domains, the XIAP protein is essential for inhibiting apoptosis. The XIAP-NFT and XIAP-VTR complexes that were produced by docking were each subjected to 10 ns MD simulations in this investigation after NFT had been docked into the XIAP. It was then determined that peptides bound free energy to XIAP using the MM/PBSA technique. The outcomes of the RMSD and RSMF methods matched the docking and previous experimental results quite well.
According to the findings, NFT and VTR compounds were the most effective P. crocatum compounds in terms of binding free energy to XIAP. Additionally, analysis of the bonds forming these NFT-XIAP domains in the final complex structures revealed that the BIR3 domain’s Leu 307, Thr 308, Glu 314, and Tyr 324 were necessary for peptide binding. The MD simulation’s energy decomposition results for attaching this NFT to the XIAP domain were in line with earlier findings and supported the functions of the same residues. The presence of an aromatic group in position four and a positive charge in position two of these peptides, when compared to other compounds, resulted in a greater affinity for binding as compared to the native ligand.
Although overt host cell death can result in tissue damage, host cell death is an essential physiological mechanism for promoting an organism’s health. A natural cell death inhibitor called XIAP blocks the action of several caspase enzymes. X-linked lymphoproliferative disease-2 is a human condition caused by inactivating mutations of XIAP that are characterized by immunodeficiency and lymphoproliferation; the underlying processes of this condition remain unclear. Through distinct impacts on the innate and acquired immune systems, we have examined the function of XIAP in innate and acquired immune responses against two intracellular bacteria in this article. The results demonstrate that XIAP supports the development of acquired immune responses.
Piper crocatum compounds may inhibit CA through multiple mechanisms, including direct antiviral activity, induction of apoptosis via XIAP inhibition, modulation of the immune system, anti-inflammatory effects, and regulation of cell proliferation. These combined actions can effectively reduce the growth and spread of HPV-infected cells, providing a potential therapeutic approach for managing CA (Fig. 9).
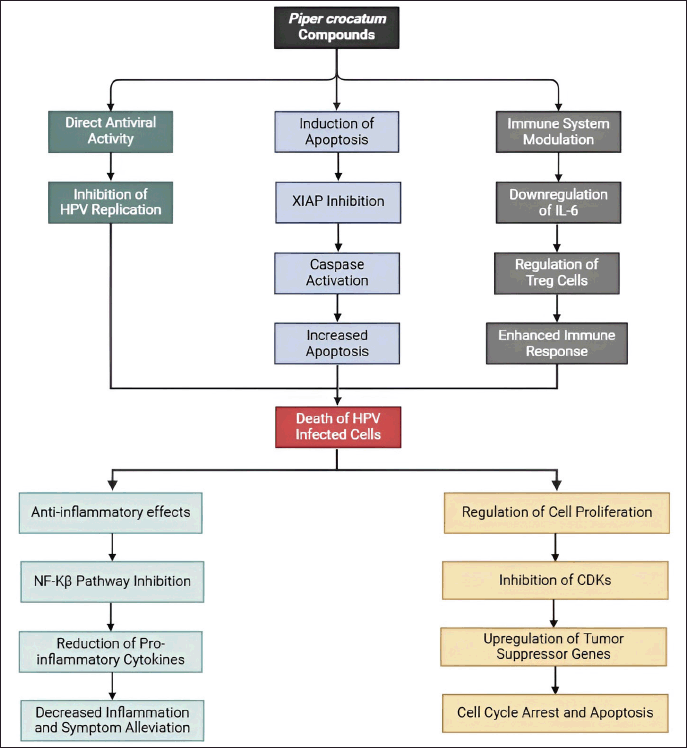 | Figure 9. Propose mechanism P. crocatum as inhibitor of CA. Color notes: green, antiviral activity; blue, induction of apoptosis; grey, immune system modulation; light blue, anti-inflammatory effects; orange, regulation of cell proliferation. [Click here to view] |
However, there is limited data availability presented in this study. The availability of comprehensive datasets on the molecular interactions and pathways involved in P. crocatum’s effects on CA may be limited, which could constrain the depth of the network analysis. Complexity of Network Interactions: Molecular interactions involved in the inhibition of CA are likely to be complex and multifaceted. Capturing this complexity in a network analysis may be challenging.
CONCLUSION
The enriched pathways’ alignment with the established biology of the CA mechanism is taken into consideration, as is the possibility that they provide new insights into the relationship between the XIAP-targeted intrinsic pathway for apoptosis, the immune system, and cytokine signaling. These simulations facilitate the study of protein-protein interactions. As strong interaction molecules, these two phytochemicals stabilized with the least amount of displacement from the interaction site during the observed entire simulation time, as confirmed by the docking and MD simulation. Furthermore, to assess the binding stability of these plant chemical complexes with XIAP proteins, protein docking was performed. The outcomes demonstrated that they could lessen XIAP proteins’ capacity to bind to NFT. Consequently, these phytochemicals might be good candidate medications to treat CA. To ascertain their efficacy as antiviral medicines, a number of comprehensive preclinical investigations are required.
ACKNOWLEDGMENTS
The author’s gratitude to all colleagues from the Laboratory of Department of Chemistry, Faculty of Medicine, Universitas Indonesia, Jakarta, who participated in this study.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
This research was supported by the RIIM LPDP Grant and BRIN, grant number (184/IV/KS/11/2023 and 04313/UN4.22/PT.01.03/2023).
CONFLICT OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
The original data presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Henri K, Eljona X, Frenki V, Asfloral H, Bledi H, Eriol B, et al. Surgical treatment of a severe anogenital condyloma acuminata—case report. J Educ Pract. 2023;14(16):1.
2. Peixoto A, Silva M, Castro R, Coelho R, Sarmento A, Macedo G. Anal condylomas: predictors of recurrence and progression to high-grade dysplasia/carcinoma in situ. J Gastrointest Oncol. 2017;8(6):1114–5.
3. Calik J, Zawada T, Bove T. Treatment of condylomata acuminata using a new non-vapor-generating focused ultrasound method following imiquimod 5% cream. Case Rep Dermatol. 2022;14(3):275–82.
4. Rego CMA, Francisco AF, Boeno CN, Paloschi MV, Lopes JA, Silva MDS, et al. Inflammasome NLRP3 activation induced by convulxin, a C-type lectin-like isolated from Crotalus durissus terrificus snake venom. Sci Rep. 2022;12(1):4706.
5. Mesquita KDSM, Feitosa BS, Cruz JN, Ferreira OO, Franco CJP, Cascaes MM, et al. Chemical composition and preliminary toxicity evaluation of the essential oil from Peperomia circinnata link var. circinnata. (Piperaceae) in Artemia salina leach. Mol Basel Switz. 2021;26(23):7359.
6. Santana de Oliveira M, Pereira da Silva VM, Cantão Freitas L, Gomes Silva S, Nevez Cruz J, de Aguiar Andrade EH. Extraction yield, chemical composition, preliminary toxicity of Bignonia nocturna (Bignoniaceae) essential oil and in silico evaluation of the interaction. Chem Biodivers. 2021;18(4):e2000982.
7. Costa EB, Silva RC, Espejo-Román JM, Neto MFDA, Cruz JN, Leite FHA, et al. Chemometric methods in antimalarial drug design from 1,2,4,5-tetraoxanes analogues. SAR QSAR Environ Res. 2020;31(9):677–95.
8. Zhong Y, Wei J, Song W, Wang Q, Zhang Z, Liu H, et al. Identification of novel biomarkers and key pathways of condyloma acuminata. Genomics. 2022;114(2):110303.
9. Liu D, Wang X, Du Z, Liu J, Zhong Y, Tian Q. The binding affinity prediction of PI3K/Akt/mTOR signaling pathway proteins with drugs based on deep learning method. Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); [cited 2024 May 13]. pp. 1667–72. Houston, TX: IEEE; 2021. Available from: https://ieeexplore.ieee.org/document/9669786
10. Sagerman PM, Kadish AS, Niedt GW. Condyloma acuminatum with superficial spirochetosis simulating condyloma latum. Am J Dermatopathol. 1993;15(2):176–9.
11. Li H, Yi T, Zhao S, Chen P, Cheng C, Wei Y, et al. The anti-condyloma acuminatum effects of interferon-inducible protein 10 in vitro. Int J Dermatol. 2009;48(2):136–41.
12. Astuti PDY, Fadilah F, Promsai S, Bahtiar A. Integrating molecular docking and molecular dynamics simulations to evaluate active compounds of Hibiscus schizopetalus for obesity. J Appl Pharm Sci. 2024;14(4):176–87.
13. Elcock AH, Sept D, McCammon JA. Computer simulation of protein−protein interactions. J Phys Chem B. 2001;105(8):1504–18.
14. GeneCards. Condyloma acuminata related genes—GeneCards search results [Internet]. [cited 2024 May 16]. Available from: https://www.genecards.org/Search/Keyword?queryString=condyloma%20acuminata
15. STRING. Functional protein association networks [Internet]. [cited 2024 May 16]. Available from: https://string-db.org/
16. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(Web Server issue):W90–7.
17. Bank RPD. RCSB PDB—5OQW: XIAP in complex with small molecule [Internet]. [cited 2024 May 16]. Available from: https://www.rcsb.org/structure/5OQW
18. Marvin. The only chemical drawing tool you will ever need [Internet]. [cited 2024 May 16]. Available from: https://chemaxon.com/marvin
19. Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, et al. SwissDock 2024: major enhancements for small-molecule docking with attracting cavities and AutoDock Vina. Nucleic Acids Res. 2024 Apr 30;gkae300.
20. YASARA. Yet another scientific artificial reality application [Internet]. [cited 2024 May 16]. Available from: https://www.yasara.org/
21. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–64.
22. PharmMapper [Internet]. [cited 2024 May 16]. Available from: https://www.lilab-ecust.cn/pharmmapper/
23. KEGG PATHWAY. KEGG PATHWAY Database [Internet]. [cited 2024 May 16]. Available from: https://www.genome.jp/kegg/pathway.html
24. Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, et al. SwissDock 2024: major enhancements for small-molecule docking with attracting cavities and AutoDock vina. Nucleic Acids Res. 2024;2024:gkae300.
25. Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9(10):679–91.
26. Ma W, Melief CJ, van der Burg SH. Control of immune escaped human papilloma virus is regained after therapeutic vaccination. Curr Opin Virol. 2017;23:16–22.
27. Pahne-Zeppenfeld J, Schröer N, Walch-Rückheim B, Oldak M, Gorter A, Hegde S, et al. Cervical cancer cell-derived interleukin-6 impairs CCR7-dependent migration of MMP-9-expressing dendritic cells. Int J Cancer. 2014;134(9):2061–73.
28. Saeki H, Moore AM, Brown MJ, Hwang ST. Cutting edge: secondary lymphoid-tissue chemokine (SLC) and CC chemokine receptor 7 (CCR7) participate in the emigration pathway of mature dendritic cells from the skin to regional lymph nodes. J Immunol. 1999;162(5):2472–5.
29. Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8(+) T cells after infection. Nat Immunol. 2002;3(7):619–26.
30. Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516.
31. McComb S, Mulligan R, Sad S. Caspase-3 is transiently activated without cell death during early antigen driven expansion of CD8+ T cells in vivo. PLoS One. 2010;5(12):e15328.
32. Ch’en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM. Mechanisms of necroptosis in T cells. J Exp Med. 2011;208(4):633–41.
33. Feng Y, Daley-Bauer LP, Roback L, Guo H, Koehler HS, Potempa M, et al. Caspase-8 restricts antiviral CD8 T cell hyperaccumulation. Proc Natl Acad Sci U S A. 2019;116(30):15170–7.
34. Deveraux QL, Reed JC. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;13(3):239–52.
35. Lopes RB, Gangeswaran R, McNeish IA, Wang Y, Lemoine NR. Expression of the IAP protein family is dysregulated in pancreatic cancer cells and is important for resistance to chemotherapy. Int J Cancer. 2007;120(11):2344–52.
36. Obexer P, Ausserlechner MJ. X-linked inhibitor of apoptosis protein—a critical death resistance regulator and therapeutic target for personalized cancer therapy. Front Oncol. 2014;4:197.
37. Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24(3):645–55.
38. Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11(2):519–27.
39. Yin GW, Guo Y, Huang YH, Song FJ. The expression and significance of XIAP and C-jun on condyloma acuminatum. Pak J Pharm Sci. 2015;28(4 Suppl.):1551–3.
40. Stuqui B, Provazzi PJS, Lima MLD, Cabral ÁS, Leonel ECR, Candido NM, et al. Condyloma acuminata: an evaluation of the immune response at cellular and molecular levels. PLoS One. 2023;18(4):e0284296.
41. Wu S, Chen H. Anti-condyloma acuminata mechanism of microRNAs-375 modulates HPV in cervical cancer cells via the UBE3A and IGF-1R pathway. Oncol Lett. 2018;16(3):3241–7.
42. Shi YJ, Yang J, Yang W. Mechanistic investigation of immunosuppression in patients with condyloma acuminata. Mol Med Rep. 2013;8(2):480–6.
43. Yang Z, Xiong H, Wei S, Liu Q, Gao Y, Liu L, et al. Yes-associated protein promotes the development of condyloma acuminatum through EGFR pathway activation. Dermatol Basel Switz. 2020;236(5):454–66.