INTRODUCTION
Lung cancer is the deadliest cancer because it is one of the most chronic types of cancer in the world [1–3] and can occur in men and women [4,5]. This disease based on basal epithelial cells is classified into two types, namely non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) [6]. NSCLC accounts for around 85% of lung cancer cases, with details of 40% adenocarcinoma, 5%–10% large cell carcinoma, and the remaining 30% originating from squamous cells. Meanwhile, SCLC will account for 15% of all cancers consisting of normal, undifferentiated small cells by 2030 [3,7].
Lung cancer could be caused by tobacco smoking, environmental exposures (such as second-hand smoke, air pollution, and radon), exposure to asbestos and arsenic, and epigenetic changes [8]. Commonly used treatments for lung cancer include surgery, radiation therapy, chemotherapy, molecular targeted therapy including epidermal growth factor receptors or anaplastic lymphoma kinase inhibition, and immunotherapy [9,10]. Although advanced treatment methods have been developed, the prognosis for lung cancer remains very unsatisfactory. Recent research reports that chemical compounds extracted from natural products are potential and effective for the treatment of lung cancer. Drug formulations from natural ingredients, namely paclitaxel, doxorubicin, and camptothecin, have been reported to have high efficacy in the treatment of lung cancer [11]. Therefore, targeted cancer therapy with specific signaling using natural compounds is promising and is being developed into pre-clinical and clinical trials [12]. Specifically, treatment with various combinations of natural compounds shows good effectiveness and safety levels in the treatment of lung cancer, which were tested in vivo [13], so they are promising in the development of bioactive compounds derived from natural ingredients.
The royleanone compounds and their derivatives from natural materials have been successfully isolated, including 7α-acetoxyroyleanone and Horminone from the Peltodon longipes plant, and showed a cytotoxic effect on pancreatic cancer cells (MIAPaca-2) and melanoma cancer cells (MV-3) [14,15]. Hormone compounds from the plant Salvia lachnocalyxdan show cytotoxicity against breast cancer cells (MCF-7) and human chronic myelogenous leukemia (K-562) [16]. The compounds 16-hydroxy-7α-acetoxyroyleanone and 16-acetoxy-7α-hydroxyroyleanone were successfully isolated from the leaves of the Coleus amboinicus, Lour., plant and showed potential as antioxidants [17,18]. The chemical structure of 16-hydroxy-7α-acetoxyroyleanone and 16-acetoxy-7α-hydroxyroyleanone is shown in Figure 1. These compounds belong to royleanone derivatives, which are reported to have potential in cancer treatment. Based on the previous information, we will continue to study the potential application of both royleanone derivatives in the treatment of lung cancer using bioinformatic approaches and network pharmacology as initial assays before further developing in vitro and in vivo tests.
MATERIALS AND METHODS
Materials
In general, the tools used in molecular docking include computers (Intel Core I5-10400F, 16 GB RAM DDR4, 256GB SSD NVME, and VGA AMD Radeon RX6600) with a Windows 10 Home operating system, Auto Dock Tools 1.5.6, PyRx 0.8, PyMOL 2.3, GaussView 5.0 (MDL Information Systems, Inc.), and Discovery Studio 21.0 Client (DSV 19.0) software. The natural compounds that were studied in this study through a bioinformatic approach were 16-hydroxy-7-acetoxyroyleanone and 16-acetoxy-7α-hydroxyroyleanone. The bioinformatics approach to the isolated compounds used the Swiss target prediction database, Cytoscape 3.10.1, Online Mendelian Inheritance in Man (OMIM), DisGeneNet, therapeutic targets database [TTD], and GeneCards.
Isolated active compound
Two active compounds, named 16-hydroxy-7α-acetoxyroyleanone and 16-acetoxy-7α-hydroxyroyleanone were isolated from the ethyl acetate extract of the leaves of the C. amboinicus, Lour., plant. The ethyl acetate extract was obtained from the multilevel partitioning of the methanol extract with n-hexane, chloroform, and ethyl acetate, as reported earlier [17,18]. Structures of isolated compounds were built with GaussView 5.0 (MDL Information Systems, Inc.) and optimized with Gaussian 09W using the density function theory (DFT) method with the B3LYP hybrid function and 3-21G(d,p) basis set [19].
Identification of the potential activity of active compounds against lung cancer
Active compounds resulting from 3-D optimization were opened with ChemDraw to obtain SMILE data for predicting target genes and molecular docking. SMILE active compound structure data was uploaded to the Swiss target prediction database [http://www.swisstargetprediction.ch/]. The predicted target gene data was downloaded in CSV format, and the data was filtered and integrated using Microsoft Excel software. Predicted target components were imported to UniProt for the normalization process, followed by the restriction to the human species. Afterward, all target proteins were retrieved and corrected according to common names. Anticancer lung targets of the active compounds were imported into Cytoscape (3.10.1) to generate “target-active compounds” tissue. The link between the active compound and the target is called a node, and the correlation between the active compound and the target is called an edge. Information on proteins involved in lung cancer was obtained using the National Center for Biotechnology Information GeneCards database [https://www.genecards.org/], TTD (https://db.idrblab.net/ttd/), OMIM (https://www.omim.org/), and DisGeNET using the keyword “lung cancer.” Both information on active compound components and treatment target components in lung cancer were processed using VENNY 2,1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html) to obtain Venn diagrams of active compounds and with lung cancer as the component targets [12,20,21].
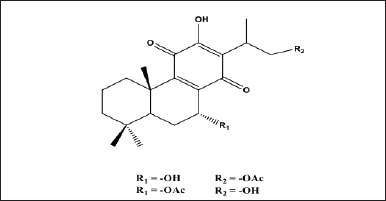 | Figure 1. Royleanone compounds isolated from the ethyl acetate extract of C. amboinicus, Lour., leaves. [Click here to view] |
Protein-protein interaction (PPI) analysis and screening
PPI determines the interaction relationship between two or more proteins based on biochemistry, hydrophobicity, and electrostatics. Protein is an important part of biological processes that occur in living things, both in healthy and diseased conditions. Gene and protein interconnections are represented by PPI, as this network is an important part of bioinformatics research [20]. The protein that intersects the active compound and lung cancer is uploaded to the STRING online site (https://string-db.org/) with the settings Homo sapiens with a confidence level of 0.4, and other parameters were selected as default. PPI relationships with node1, node2, and the combined score from the export file are imported into Cytoscape 3.10.1 to form an interaction network. The results of the analysis of the top target were carried out using the CytoHubba plugin to obtain the highest protein with the highest 20-degree value for the next stage [12,21].
Gene Ontology (GO) and Kyoto Encyclopedia of Genes, and Genomes (KEGGs) pathway analysis
GO and KEGG analyses were performed with the approach of biological processes, cellular components, gene functionality, cellular components, and molecular functions. The results of bioinformatic analysis of STRING data [12,21] were obtained by importing active compounds intercept targets with lung cancer to result in the molecular mechanisms of active compounds in the treatment of lung cancer.
Molecular docking approach
Molecular docking is a rational drug design method based on exploring the interaction between ligands (active compounds) and receptors (proteins). The interactions were studied to predict the binding affinity of active compounds for proteins, as well as the intermolecular patterns conferring biological activity [22,23]. Molecular docking could be carried out using AutoDock Vina, developed by the Scripps Research Institute, for semi-flexible molecular docking computing. AutoDock Vina uses complex gradient algorithms and multi-threaded techniques to produce predictions that are more accurate and faster when compared to AutoDock 4 [21]. The optimized active compounds were tethered to three cancer proteins obtained from the KEGG cancer pathway. The protein codes were traced to the PDB database [https://www.rcsb.org/] and selected specifically for lung cancer. The best protein crystal structures were selected, and the PDB database was downloaded. Protein PDB files of active compounds and ligand molecules are imported into the AutoDock tool for molecular docking. AutoDock Vina was run to bind the active compound treated to the target protein ten times, and the lowest binding energy for each docking was taken as the final result. Complexes were then observed and plotted using PyMOL [21].
RESULTS AND DISCUSSION
Optimization of active compounds
Two active compounds, namely 16-hydroxyroyleanone-7α-acetoxyroyleanone and 16-acetoxy-7α-hydroxyroyleanone, were prepared by optimizing their structure to obtain the most stable energy with Gaussian 09W using the DFT method with B3LYP hybrid function and 6-31G(d,p) basis set. Their structures are then validated with AutoDockTools, as shown in Table 1. The optimizing process of the compounds was carried out to obtain thermal stability and molecular charge as well as their biochemical behavior based on quantum mechanics calculations [24]. The results of molecular optimization are expected to have the same molecular structure in the human body.
Identification of active compounds against lung cancer
The Swiss Target Prediction results had the densest 50 targets from 16-acetoxy-7α-hydroxyroyleanone (Compound 1) and 54 targets from 16-hydroxy-7α-acetoxyroyleanone (Compound 2). At the same time, the combined prediction of both active compounds produced 104 targets. Predictions of 104 total targets were analyzed by eliminating targets so they were not doubled to obtain a total of 77 targets. The resulting active compound-target network was formed using Cytoscape 3.10.0 software (Fig. 2a). Based on validation results from various databases, 18,404 protein targets associated with lung cancer were identified. Both 16-acetoxy-7α-hydroxyroyleanone and 16-hydroxyroyleanone-7α-acetoxyroyleanone have intercepted 77 potential targets that are predicted to be related to lung cancer proteins (Fig. 2b).
Target analysis of PPI networks
The total targets predicted (77 targets) that had intercepts with lung cancer were analyzed and then imported into a string to build a PPI network by selecting Homo sapiens organisms with a confidence of 0.40 (Fig. 3a). Intercepts of active target-compounds were imported into Cytoscape 3.10.0 to construct and obtain a network map of potential target interactions (Fig. 3b). The results of the analysis showed that there were 77 nodes (proteins) and 214 edges (interactions). Based on the linkages informing the PPI, the 10 highest (highest to lowest) interaction targets include prostaglandin-endoperoxide synthase-2 (PTGS2), peroxisome proliferator-activated receptor gamma (PPARG), ACE, MMP2, ITGB1, SERPINE1, REN, ITGB3, PPARA, KDR and they were considered to have the best relationship prediction for lung cancer.
 | Table 1. The active compounds isolated from the ethyl acetate extract of C. amboinicus, Lour leaves. [Click here to view] |
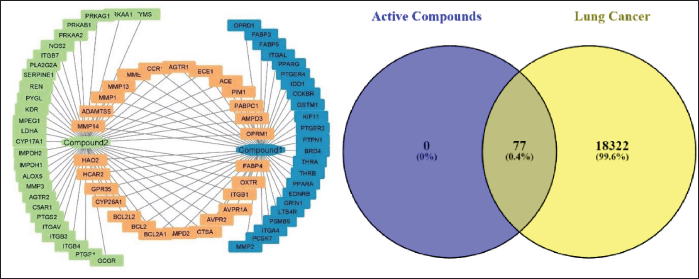 | Figure 2. Identification of active-target compounds: (a) Active-target combined network with Cytoscape 3.10.0 and (b) intercept Venn diagram of each compound against lung cancer targets. [Click here to view] |
 | Figure 3. Analysis of PPI: (a) PPI network built from intercept prediction of active-target compounds using strings, and (b) proteins (nodes) that are linked by interactions (edges) with one another, and blue color represents the highest PPI top 10.ç [Click here to view] |
GO and the KEGG pathway analysis
GO and KEGG pathways were analyzed to explore the mechanisms of 77 targets in the treatment of lung cancer using a bioinformatics database [ShinyGO 0.77]. GO and KEGG results of active compounds against lung cancer are displayed in Figure 4. The analysis is carried out on biological processes, cellular components, molecular functions, and the KEGG pathway of the potential targets of active compounds in the treatment of lung cancer, which are shown in Figure 4a–d, respectively.
The enrichment results identified the 20 pathways associated with active compound targets in the treatment of lung cancer that have been analyzed according to the KEG pathways from potential targets of active compounds. The targets associated with each of these pathways are described in Table 2. Based on the cancer pathway, there are 14 targets related to the cancer pathway, i.e., MMP2, PTGER2, ITGAV, PPARG, PTGER4, GSTM1, MMP1, NOS2, PTGS2, PIM1, EDNRB, ITGB1, BCL2, and AGTR1. The two targets with the highest interaction between active compounds and lung cancer in the cancer pathway are found in PTGS2 and PPARG.
In silico and molecular docking
The two best target interactions of active compounds for the treatment of lung cancer are selected based on the results of KEGG pathways analysis, namely PTGS2 and PPARG. The PTGS2 plays an important pathological role in the treatment of chemotherapy-resistant cancer. Moreover, PTGS2 has a role in increasing the response of bodies that are resistant to chemotherapy treatment [25,26]. Therapy for non-drug-resistant cancer cells could be done by inhibiting the expression of PTGS2. Inhibition of PTGS2 expression will suppress the proliferation, migration, and invasion of cancer cells, as well as modulate the immune response by impairing cell differentiation and suppressing the occurrence of metastases [27]. On the other hand, the PPARG target is the most abundant subtype expressed in adipose tissue with two isoforms. The inhibition of the PPARG target is able to inhibit cell proliferation, induce cell cycle arrest, apoptosis of multiple cancer cells, increase adhesion between cells, and immobilize the tumor microcell environment, which promotes both transcription and protein levels and prevents metastases [28,29]. In addition, PPARG also plays an important role in regulating the expression of various genes regarding glucose and lipid metabolic homeostasis, adipogenesis, and inflammation [30]. These two targets of active compounds for the treatment of lung cancer have two opposite directions: PPARG inhibits apoptosis, signaling, proliferation, and metastases, thus overcoming the occurrence of resistance and increasing the activity of cancer treatment [31,32].
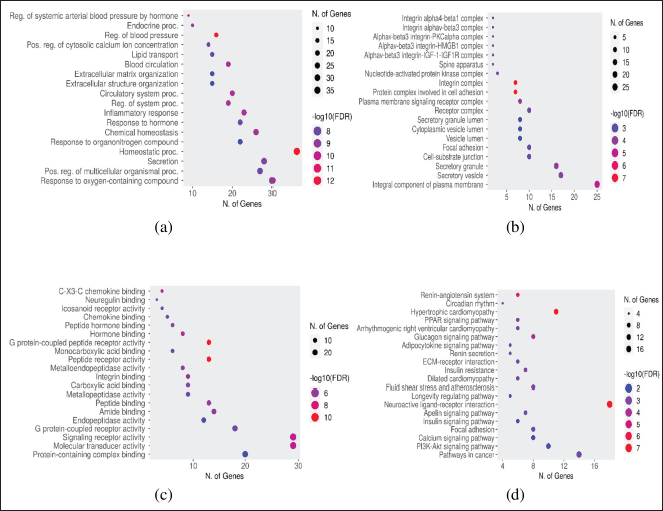 | Figure 4. GO and KEG analysis of active compounds for the treatment of lung cancer was built using the ShinyGO 0.77 database: [a] Biological processes, (b) cellular components, (c) molecular functions, and (d) KEGG pathways from potential targets of active compounds in the treatment of lung cancer. [Click here to view] |
 | Table 2. Pathways related to active compounds and lung cancer. [Click here to view] |
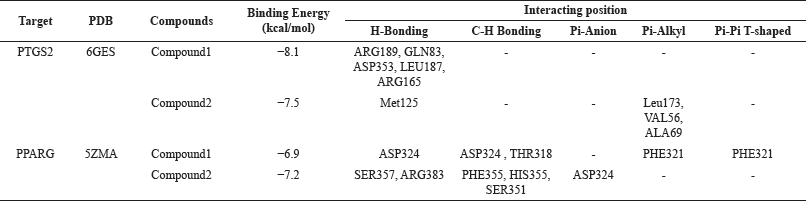 | Table 3. Strength of interaction between active compounds and potential lung cancer target proteins. [Click here to view] |
Besides, suppressing or inhibiting the expression of the PTGS2 target resulted in a maximum treatment and inhibition of cancer invasion and metastases [31,33]. In silico testing of the active compounds was carried out on target proteins that have been reported as a therapy in cancer treatment. The protein codes used in this approach are 6GES (PTGS2 protein) and 5ZMA (PPARG protein) (Table 3 and Fig. 5).
 | Figure 5. 2-D interactions of molecular docking of active compounds against lung cancer target proteins: (a) 16-acetoxy-7α-hydroxyroyleanone and PTGS2 receptor, (b) 16-hydroxy-7α-acetoxyroyleanone and PTGS2 receptor, (c) 16-acetoxy-7α-hydroxyroyleanone and PPARG receptor, and (d) 16-hydroxy-7α-acetoxyroyleanone and PPARG receptor. [Click here to view] |
CONCLUSION
The reported bioinformatic study of two compounds isolated from the leaves of C. amboinicus Lour., namely 16-acetoxy-7α-hydroxyroyleanone and 16-hydroxy-7α-acetoxyroyleanone, suggests potential application in the treatment of lung cancer. The results of integrated bioinformatic analysis of GO, KEGG, and cancer pathways have 77 targets, with PTGS2 and PPARG being the two main targets for treating lung disease. Validated molecular docking analysis revealed multiple beneficial interactions in the active sites of PTGS2 and PPARG receptors. These findings are important as initial information and need to be developed into further research stages. This research is still very limited because it was only carried out based on bioinformatic studies, so further testing needs to be done in vitro and in vivo regarding these two active compounds as therapeutic targets in the treatment of lung cancer.
ACKNOWLEDGMENT
The authors are grateful for the support of facilities from the Faculty of Mathematics and Natural Sciences, Universitas Gadjah Mada, Yogyakarta, while conducting this research
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Carter BW, Halpenny DF, Ginsberg MS, Papadimitrakopoulou VA, De Groot PM. Immunotherapy in non-small cell lung cancer treatment: current status and the role of imaging. J Thorac Imaging. 2017;32(5):300–12. CrossRef
2. Islami F, Torre LA, Jemal A. Global trends of lung cancer mortality and smoking prevalence. Transl Lung Cancer Res. 2015;4(4):327–38. CrossRef
3. Sharma P, Mehta M, Dhanjal DS, Kaur S, Gupta G, Singh H, et al. Emerging trends in the novel drug delivery approaches for the treatment of lung cancer. Chem Bio Interact. 2019;25:108720. CrossRef
4. Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24. CrossRef
5. Majeed U, Manochakian R, Zhao Y, Lou Y. Targeted therapy in advanced non-small cell lung cancer: current advances and future trends. J Hematol Oncol. 2021;14(1):108. CrossRef
6. Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The effect of advances in lung-cancer treatment on population mortality. N Engl J Med. 2020;383(7):640–9. CrossRef
7. Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer. 2015;90(2):121–7. CrossRef
8. Norouzi M, Hardy P. Clinical applications of nanomedicines in lung cancer treatment. Acta Biomater. 2021;121:134–42. CrossRef
9. Otsuki Y, Saya H, Arima Y. Prospects for new lung cancer treatments that target EMT signaling. Dev Dyn. 2018;247:462–72. CrossRef
10. Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers. 2021;7(1):1–20. CrossRef
11. Wen T, Song L, Hua S. Perspectives and controversies regarding the use of natural products for the treatment of lung cancer. Cancer Med. 2021;10(7):2396–422. CrossRef
12. Iksen, Pothongsrisit S, Pongrakhananon V. Targeting the PI3K/AKT/mTOR signaling pathway in lung cancer: an update regarding potential drugs and natural products. Molecules. 2021;26(13):1–27. CrossRef
13. Jin X, Yang Q, Cai N, Zhang Z. A cocktail of betulinic acid, parthenolide, honokiol and ginsenoside Rh2 in liposome systems for lung cancer treatment. Nanomedicine. 2019;15(1):41–54. CrossRef
14. Fronza M, Lamy E, Günther S, Heinzmann B, Laufer S, Merfort I. Abietane diterpenes induce cytotoxic effects in human pancreatic cancer cell line MIA PaCa-2 through different modes of action. Phytochemistry. 2012;78:107–19. CrossRef
15. Fronza M, Murillo R, ?lusarczyk S, Adams M, Hamburger M, Heinzmann B, et al. In vitro cytotoxic activity of abietane diterpenes from Peltodon longipes as well as Salvia miltiorrhiza and Salvia sahendica. Bioorg Med Chem. 2011;19(16):4876–81. CrossRef
16. Mirzaei HH, Firuzi O, Jassbi AR. Diterpenoids from roots of Salvia lachnocalyx; in-silico and in-vitro toxicity against human cancer cell lines. Iran J Pharm Res. 2020;19(4):85–94. CrossRef
17. Gurning K, Haryadi W, Sastrohamidjojo H. Isolation and characterization of antioxidant compounds of Bangun-Bangun (Coleus amboinicus, l.) leaves from North Sumatera, Indonesia. Rasayan J Chem. 2021;14(1):248–53. CrossRef
18. Gurning K, Haryadi W. Potential antioxidants of secondary metabolite isolates ethyl acetate fraction Coleus amboinicus Lour. Leaves. ScienceRise: Pharm Sci. 2022;39(5):100–5. CrossRef
19. Haryadi W, Pranowo HD. Molecular docking and dynamics analysis of halogenated imidazole chalcone as anticancer compounds. Pharmacia. 2023;70(2):323–9. CrossRef
20. Basar MA, Hosen MF, Kumar Paul B, Hasan MR, Shamim SM, Bhuyian T. Identification of drug and protein-protein interaction network among stress and depression: a bioinformatics approach. Inform Med Unlocked. 2023;37:1–10. CrossRef
21. Wang Y, Zhang Y, Wang Y, Shu X, Lu C, Shao S, et al. Using network pharmacology and molecular docking to explore the mechanism of Shan Ci Gu (Cremastra appendiculata) against non-small cell lung cancer. Front Chem. 2021;9:1–14. CrossRef
22. Vidal-Limon A, Aguilar-Toalá JE, Liceaga AM. Integration of molecular docking analysis and molecular dynamics simulations for studying food proteins and bioactive peptides. J Agric Food Chem. 2022;70(4):934–43. CrossRef
23. Li T, Guo R, Zong Q, Ling G. Application of molecular docking in elaborating molecular mechanisms and interactions of supramolecular cyclodextrin. Carbohydr Polym. 2022;276:1–15. CrossRef
24. Rana KM, Maowa J, Alam A, Dey S, Hosen A, Hasan I, et al. In silico DFT study, molecular docking, and ADMET predictions of cytidine analogs with antimicrobial and anticancer properties. In Silico Pharmacol. 2021;9(42):1–24. CrossRef
25. Bell CR, Pelly VS, Moeini A, Chiang SC, Flanagan E, Bromley CP, et al. Chemotherapy-induced COX-2 upregulation by cancer cells defines their inflammatory properties and limits the efficacy of chemoimmunotherapy combinations. Nat Commun. 2022;13(1):1–17. CrossRef
26. Lin XM, Luo W, Wang H, Li RZ, Huang YS, Chen LK, et al. The role of prostaglandin-endoperoxide synthase-2 in chemoresistance of non-small cell lung cancer. Front Pharmacol. 2019;10(836):1–14. CrossRef
27. Ercolano G, De Cicco P, Rubino V, Terrazzano G, Ruggiero G, Carriero R, et al. Knockdown of PTGS2 by CRISPR/CAS9 system designates a new potential gene target for melanoma treatment. Front Pharmacol. 2019;10(1456):1–12. CrossRef
28. Ravi Kiran Ammu VVV, Garikapati KK, Krishnamurthy PT, Chintamaneni PK, Pindiprolu SKSS. Possible role of PPAR-γ and COX-2 receptor modulators in the treatment of non-small cell lung carcinoma. Med Hypotheses. 2019;124:98–100. CrossRef
29. Shi S, Yu G, Huang B, Mi Y, Kang Y, Simon JP. PPARG could work as a valid therapeutic strategy for the treatment of lung squamous cell carcinoma. PPAR Res. 2020;1:1–9. CrossRef
30. Tan Y, Wang M, Yang K, Chi T, Liao Z, Wei P. PPAR-α modulators as current and potential cancer treatments. Front Oncol. 2021;11:1–15. CrossRef
31. Liang X, Wang J, Liu Y, Wei L, Tian F, Sun J, et al. Polymorphisms of COX/PEG2 pathway-related genes are associated with the risk of lung cancer: a case–control study in China. Int Immunopharmacol. 2022;108:1–7. CrossRef
32. S?owikowski BK, Drzewiecka H, Malesza M, M?dry I, Sterzy?ska K, Jagodzi?ski PP. The influence of conjugated linoleic acid on the expression of peroxisome proliferator-activated receptor-γ and selected apoptotic genes in non-small cell lung cancer. Mol Cell Biochem. 2020;466(1–2):65–82. CrossRef
33. Cancemi P, Buttacavoli M, Roz E, Feo S. Expression of alpha-enolase (ENO1), Myc promoter-binding protein-1 (MBP-1) and matrix metalloproteinases (MMP-2 and MMP-9) reflect rhe nature and aggressiveness of breast tumors. Int J Mol Sci. 2019;20(16):3952. CrossRef