INTRODUCTION
Chemotherapy is a widely used treatment modality for cancer. However, its use is challenging because it damages both cancerous and healthy tissues [1]. Since the type, dose, and duration of chemotherapy critically influence patient health and treatment outcomes [2], there is an increasing focus on exploring adjunct therapies that can enhance cancer patients’ health while imposing fewer side effects [3]. Etoposide is a chemotherapeutic drug for the treatment of various malignancies, including small-cell lung cancer, testicular cancer, and lymphomas [4]. In addition, etoposide is commonly used to treat Hodgkin’s lymphoma and AIDS as well as ovarian, uterine, bladder, and prostate cancers [5,6]. Etoposide exerts anticancer effects by inhibiting topoisomerase II, a key enzyme in DNA replication and repair [7]. Specifically, etoposide inhibits the re-ligation step of the enzyme’s catalytic cycle, causing the accumulation of DNA double-strand breaks. This DNA damage triggers apoptosis in rapidly dividing cancer cells [8]. Several studies report that etoposide frequently elevates liver enzymes, and high doses can induce acute liver injury, including sinusoidal obstruction syndrome [9]. Furthermore, after receiving treatment for 1–5 months, etoposide has been connected to occurrences of acute hepatitis that have often been mild but can occasionally become severe [10]. It has been shown that topoisomerase II inhibitors like etoposide are known to undergo substantial metabolism in the liver, and this process has been linked to significant hepatocellular damage. Etoposide treatment has been associated with a notable increase in serum levels of alkaline phosphatase, aspartate aminotransferase (AST), and alanine aminotransferase (ALT) marked decrease in serum albumin levels [8]. Additionally, hepatotoxicity and immunomodulation are major adverse effects of etoposide chemotherapy [11]. Adjuvant therapies—natural or synthetic compounds—are increasingly used alongside chemotherapy to mitigate adverse effects. Biobran/MGN-3 is an arabinoxylan obtained from rice bran and altered by hydrolyzing it with shiitake mushroom enzymes. Biobran consists of arabinose monomers linked to a xylose backbone, and its molecular weight ranges from 30 to 50 kDa [12]. Several in vivo, in vitro, and human investigations suggest that biobran can enhance the performance of innate and adaptive immune cells, such as B cells, T cells, natural killer cells, dendritic cells, and macrophages [13]. It is noteworthy that previous research demonstrated that biobran could sensitize metastatic breast cancer cells to paclitaxel in vitro and enhance the apoptotic effect of a low dose of paclitaxel on tumor cells. Biobran can also make human breast cancer cells more sensitive to the chemotherapy drug daunorubicin [14]. Furthermore, biobran has chemo-preventive action against the chemical induction of glandular stomach cancer in rats [15]. Preclinical findings suggest biobran may help manage terminal hepatocellular carcinoma through increased immunity and anti-inflammatory effects, potentially improving quality of life as well as supplementing standard therapies [16].
While etoposide demonstrates efficacy as an anticancer medication, it is crucial to systematically monitor for hepatotoxicity and consider the potential for immunomodulation when incorporating it into chemotherapy protocols. Therefore, the exploration of therapeutic strategies to counteract these detrimental effects is crucial in improving patient outcomes during chemotherapy [17].
This study investigates biobran’s potential to mitigate etoposide-induced hepatotoxicity and immunosuppression, which could revolutionize adjuvant chemotherapy strategies. This combination holds promise for patients, offering a pathway to improve their condition and mitigate the adverse effects associated with chemotherapy.
MATERIALS AND METHODS
Drugs and chemicals
Etopul 100 mg/5 ml sol (etoposide) solution for infusion vial was purchased from EIMC Pharmaceuticals Company (Cairo, Egypt) (Product Code: 11204). All the other chemicals used were of the highest purity and analytical grade purchased from commercial suppliers.
Biobran/MGN-3
Biobran is a natural compound derived from the processing of rice bran with hydrolyzing enzymes sourced from Shiitake mushrooms. Its principal chemical composition features an arabinoxylan with a xylose constituent in its primary chain and an arabinose polymer in its side chain. Biobran was freshly prepared prior to each administration by dissolving it in 0.9% saline solution and was administered every other day for a duration of 6 weeks. The supply of biobran for this investigation was procured from Daiwa Pharmaceuticals Co Ltd, based in Tokyo, Japan.
Animals
A cohort of 40 healthy adult male Wistar albino rats, aged 6–7 weeks and weighing about 130 ± 5 g, were obtained from the National Research Center’s Animal House in Dokki, Cairo, Egypt. The rodents were acclimated in a specific-pathogen-free environment, housed in clean plastic cages, and provided ad libitum access to tap water and a typical diet of pellets. The animals were kept in room temperature conditions of 24°C ± 2°C, a relative humidity ranging from 60% to 70%, and a 12-hour light–dark cycle for a duration of 2 weeks preceding the commencement of the experiment. This study protocol received ethical approval under the designation (DMU-SCI-240101) from Scientific Research Ethics Committee, Faculty of Science, Damanhour University, Egypt.
Experimental design
A random classification was applied to divide 40 rats into 4 primary experimental groups, each comprising 10 rats. The groups consisted of (G1) Control Group: baseline control receiving neither etoposide nor biobran; (G2) Biobran Group: intraperitoneal injections of biobran every other day at a dosage of 40 mg/kg body weight for a period of 6 weeks [18]; (G3) Etoposide Group: intraperitoneal injections of etoposide at a dosage of 1 mg/kg body weight/day for a period of 6 weeks [19]; and (G4) Dual Group: intraperitoneal injections of both biobran at the same dosage as Group G2 and etoposide at the same dosage as Group G3.
Preparations for sample collection
Following the completion of the experimental period, rats underwent an overnight fasting period and were subsequently anesthetized using inhalant anesthesia with isoflurane.
Blood collection
Blood was collected from the tail vein using a sterile needle, with an emphasis on avoiding bone injury. The collected blood was directed into sterile tubes or microtubes for subsequent analysis, with the volume tailored to experimental requirements.
Liver tissues
Animals were dissected and the selected organ, the liver, was quickly taken from all experimental groups. To eliminate red blood cells, the dissected livers were washed in a phosphate-buffered saline (PBS) solution. Subsequently, the organs were divided into multiple parts for various analyses. For the histopathological analyses, liver pieces were fixed in 10% neutral formalin. For apoptotic markers, caspase 3, Annexin, and Bcl2 determinations, the second portions of the livers were kept at −80°C for storage. The third sections for measuring cytokines, antioxidants, and indicators of oxidative stress in tissues were frozen at −80°C.
Tissue homogenate preparation
Frozen liver tissues were homogenized in cold lysis buffer (100 mM potassium phosphate, containing 2 mM EDTA), centrifuged for 15 minutes, and stored at −20°C for further use after homogenization.
Histopathological examination
Fixed liver in 10% buffered formalin from the normal and experimental rats were dehydrated through a graded series of ethanol and embedded in paraffin according to standard procedures. Paraffin sections (5 m thick) were mounted on glass slides and used for hematoxylin and eosin stains as a routine method after [20].
A semiquantitative grading system was employed blindly by a pathologist for the assessment of liver injury [21], examining the following parameters: Hepatocyte (HC) degeneration/vacuolation –Inflammatory cell infiltration–Venous Congestion–Necrosis–Presence of bile duct proliferation. Each parameter was graded on a 0–3 scale: 0>Normal (no abnormality) – 1>Mild changes (<25% of area) – 2>Moderate (25%–50% of area) – 3>Severe (>50% of area).
Quantitative assessment of hepatic biomarkers (liver function) in rat serum
Serum AST and ALT levels were determined using standardized enzymatic assays based on the International Federation of Clinical Chemistry reference procedures. These assays monitor the oxidation of NADH to NAD+ at 340 nm [22]. The bromocresol green binding method was used to determine albumin levels [23], and the modified Jendrassik–Grof diazo method was used to determine total bilirubin [24], with the bilirubin–diazotized sulfanilic complex’s absorbance measured at 540 nm. All assays were carried out using commercially available diagnostic kits (Sigma Diagnostics, India) in a calibrated automatic biochemistry analyzer (Cobas Integra, Roche), following the manufacturer’s instructions. Each analytical run included internal quality controls and calibrators to ensure precision and accuracy.
Quantitative analysis of Immunoglobulin M (IgM) and Immunoglobulin G (IgG) levels in rat serum using ELISA assay
IgM and IgG serum concentrations were determined using MyBioSource rat-specific ELISA kits (IgM: Cat. No. MBS9135900; IgG: Cat. No. MBS261432; San Diego, CA) following the manufacturer’s instructions. Standards and serum samples were loaded onto pre-coated 96-well plates and incubated with enzyme-linked antibodies. They were washed before the addition of 3,3’,5,5’-Tetramethylbenzidine substrate and stopping the enzymatic reaction using a stop solution. Absorbance was measured at 450 nm using a microplate reader (BioTek ELx800, USA). Immunoglobulin concentrations were calculated from standard curves. Each sample was tested in duplicate, and internal quality controls ensured the reliability and reproducibility of the results [25].
Determination of oxidative stress and antioxidant biomarkers in liver tissue homogenate
Malondialdehyde (MDA), nitric oxide (NO), superoxide dismutase (SOD), catalase (CAT), and reduced glutathione (GSH) levels were determined in homogenized rat renal tissues using commercially available Abcam (Cambridge, UK) assay kits with catalog numbers ab118970, ab65328, ab65354, ab83464, and ab138881, respectively. The tissue samples were homogenized in the appropriate buffers and centrifuged to produce clear supernatants. The assays were carried out in accordance with the manufacturer’s instructions, with each analyte producing a distinct colorimetric response. Absorbance was measured with a microplate reader, and concentrations were determined using standard curves. All samples were run in duplicate with internal controls to ensure accuracy.
Determination of inflammatory status in liver tissue homogenate
The concentrations of cytokines in liver tissue homogenates from all experimental groups were measured using rat-specific ELISA kits from Boster Biological Technology (Pleasanton, CA, USA) and Biocompare (Cambridge CB4 0GJ, UK) according to the manufacturer’s procedure. IL-4 (Cat. No. EK0406), IL-6 (Cat. No. EK0412), and IL-10 (Cat. No. EK0418) were tested using Boster kits, whereas IL-8 (Cat. No. abx052099) and IL-17 (Cat. No. EKF57850) were tested using Biocompare kits. Tissues were homogenized in ice-cold PBS with protease inhibitors and then centrifuged to extract supernatants. Standards and samples were inserted in pre-coated 96-well plates, incubated with detection antibodies specific to each cytokine, and then treated with substrate. The reaction was stopped, and the absorbance was measured at 450 nm. Cytokine concentrations were calculated from standard curves, and all measurements were performed in duplicate with appropriate quality controls [26].
Immunohistochemical staining of interleukin-1 beta (IL-1β), tumor necrosis factor-alpha (TNF-α), and transforming growth factor-beta (TGF-β) in liver tissue
The specimens were fixed in 10% neutral-buffered formalin, then paraffin-embedded and cut into 5-μm sections. For the immune-histochemical study, liver sections were deparaffinized in xylene, then rehydrated through a graded series of ethanol, and antigen retrieval was performed by heating in citrate buffer (pH 6.0) at 95°C for 20 minutes. Sections were allowed to cool to room temperature and further washed in PBS, pH 7.4.
After that, the sections were incubated for 10 minutes with 3% hydrogen peroxide to block the endogenous peroxidase activity. To prevent nonspecific binding, the sections were incubated in 5% bovine serum albumin (BSA) in PBS at room temperature for 30 minutes. Later, liver sections were incubated overnight at 4°C with the respective primary antibodies, which included: Anti-IL-1β antibody: (Catalogue No. PB9025), anti-TNF alpha antibody (Catalogue No. PA1079), and anti-TGF-β 1 antibody (Catalogue No. A00019-2) BOSTER BIOLOGICAL TECHNOLOGY, 3942 B Valley Ave, Pleasanton, CA. Following the application of primary antibodies, liver sections were washed three times with PBS and incubated with a biotinylated goat anti-rabbit IgG secondary antibody suitable for each primary antibody for 30 minutes at room temperature. Then, they were incubated with streptavidin–horseradish peroxidase complex for another 30 minutes.
The sections were developed with 3,3′-diaminobenzidine as a chromogen, yielding positive staining as indicated by the brown coloration. Slides were counterstained with hematoxylin, then dehydrated and mounted. The controls were prepared by omitting the primary antibodies. Staining intensity and distribution of IL-1β, TNF-α, and TGF-β were observed under a light microscope [27,28].
Flow cytometric analysis of apoptotic markers and T cell surface molecules
Flow cytometry was applied to analyze apoptotic markers (Caspase-3, Bcl-2, and Annexin V/PI) [29], in addition to T cell surface molecules CD4, CD8, and CD3 [30] in the liver from the four experimental groups.
Approximately 100 mg of rat liver tissue was taken from each rat, washed in ice-cold PBS to drain off residual blood, and finely minced with sterile scissors. Tissue fragments were enzymatically digested in 0.2% collagenase IV (Sigma-Aldrich, USA) in PBS and incubated for 30 minutes at 37°C under gentle agitation. The digested suspension was filtered through a 70 μm nylon cell strainer to obtain a single-cell suspension. Cells were centrifuged at 300 × g for 5 minutes at 4°C. Red blood cells were lysed with red blood cells lysis buffer (BioLegend, USA) at room temperature for 2 minutes, washed, and resuspended in flow cytometry staining buffer (PBS with 1% fetal bovine serum).
For intracellular staining, cells were fixed and permeabilized first with the eBioscience Fixation/Permeabilization Kit (Thermo Fisher Scientific, USA) and blocked with 10% normal goat serum to reduce nonspecific antibody binding. Cells were stained with the following primary antibodies: rabbit anti-Caspase-3 (Cat. No. PB9188, Boster Biological Technology; 1 µg/10° cells) and mouse anti-BCL-2 (Cat. No. 13-8800, Invitrogen). After washing, cells were incubated in the dark at 4°C for 30 minutes with secondary antibodies labeled with fluorophore: Alexa Fluor® 488 goat anti-rabbit IgG (anti-Caspase-3) and Alexa Fluor® 647 goat anti-mouse IgG (anti-BCL-2).
Annexin V- and propidium iodide (PI) staining (Cat. No. 786-1544, G-Biosciences, USA) was performed to obtain early and late apoptosis as per the manufacturer’s instructions. Both staining analysis allowed the separation of viable (Annexin-/PI-), early apoptotic (Annexin+/PI-), late apoptotic (Annexin+/PI+), and necrotic (Annexin-/PI+) cells.
For the identification of liver-resident and infiltrating T cell populations, single-cell suspensions of liver tissues were labeled with fluorochrome-conjugated monoclonal antibodies against the surface markers of T cell subsets. CD4 cells had been stained with a PE-conjugated anti-mouse CD4 antibody (Clone GK1.5, Rat IgG2b, κ; Cat. No. 100407, BioLegend, USA) at 0.25 µg per million cells. For CD8, we used 200 µg/ml of Alexa Fluor® 647-conjugated anti-mouse CD8 (Clone 32-M4, Mouse IgG2a; Cat. No. sc-1177 AF647, Santa Cruz Biotechnology, USA) diluted in PBS with 0.1% gelatin. For CD3ε, FITC-conjugated anti-human/mouse CD3 (Clone UCHT1, Invitrogen™ (eBioscience), Thermo Fisher Scientific, Waltham, MA; Cat. No. AB_2043831) 5 µl/test usage. This antibody has an excitation/emission maximum of 498/517 nm and was stored at 4°C in PBS containing 0.09% sodium azide.
Antibody staining was accomplished by incubating the cells with the specified antibody for 30 minutes at 4°C in the dark. Cells were washed twice with staining buffer (PBS + 0.1% BSA) and then resuspended in the same buffer before being extracted. To determine nonspecific binding, isotype controls of each fluorochrome-conjugated antibody were used, Rat IgG2b-PE for CD4 and Mouse IgG2a-AF647 for CD8. Following staining, cells were washed twice in staining buffer before being resuspended in the same volume of buffer for flow cytometry analysis.
Data was collected using a BD Accuri™ C6 flow cytometer (BD Biosciences, USA) and analyzed with BD Accuri C6 software. A minimum of 10,000 events per sample were acquired. FSC versus SSC gating was performed initially to select viable single cells and to exclude debris and clumps of cells. Fluorescence Minus One control and isotype controls were employed for each antibody to ensure the accuracy and specificity of the gate boundaries. These controls allowed background and nonspecific fluorescence to be excluded so that only populations of true-positive cells were being quantified.
Statistical analysis
Statistical analysis was done using SPSS program (version 26). The data are calculated and presented as means associated with the standard deviation (SD). The normality was tested using Kolmogorov–Smirnov test and the data were normally distributed. The difference among the groups has been determined using one-way ANOVA (F test) and post hoc Duncan’s test. p-value less than 0.05 was considered statistically significant.
RESULTS
Liver function analysis via evaluation of liver enzymes AST and ALT and biochemical marker albumin and T. bilirubin
As shown in Table 1, ELISA assay results revealed significant differences in biochemical and physiological markers in rat plasma among various treatment groups. In G3 (etoposide), there was a substantial increase (p < 0.05) in AST and ALT levels compared to G1 (control) and G2 (biobran). However, no significant changes were observed in G2 (biobran) or G4 (dual treatment) compared to the control. Regarding albumin levels in rat plasma, the results demonstrated a slight non-significant decrease in the etoposide group (G3) compared to the control and the other two treated groups, whereas the biobran group (G2) and dual treatment group (G4) did not show any significant changes in albumin levels when compared to the control group. The analysis of total bilirubin (T. bilirubin) in rat plasma also exhibited a highly significant increase (p < 0.05) in the etoposide group when compared to the control and the other two treated groups, while there were no significant changes observed in T. bilirubin levels in the Biobran or dual treatment groups compared to the control group.
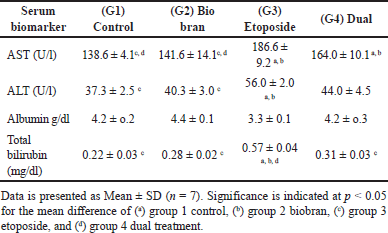 | Table 1. Shows liver enzymes AST and ALT and biochemical markers albumin and total bilirubin in rat serum across experimental groups. [Click here to view] |
Quantitative measurement of serum immunoglobulins IgM and IgG
Results in Table 2 show a significant reduction in both IgM and IgG levels in the etoposide group (G3) compared to the control and Biobran groups (p < 0.05) but not with the dual-treated group (G4). The dual treatment and biobran groups did not exhibit any significant changes in IgM and IgG levels when compared to the control group.
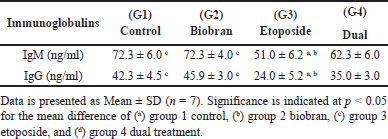 | Table 2. ELISA assay showing immunoglobulins IgM and IgG in rat serum across experimental groups. [Click here to view] |
Quantification of oxidative stress markers MDA and NO in rat liver cells homogenate from different experimental groups
The results for the oxidative stress markers MDA and NO in rat liver are depicted in Table 3. In the etoposide group (G3), there was a significant increase (p < 0.05) in both MDA and NO levels compared to the control and biobran groups. Additionally, in the dual treatment group (G4), dual treatment with biobran and etoposide decreased MDA and NO levels non-significantly compared to the etoposide group. The biobran group did not show any significant differences in MDA and NO levels compared to the control.
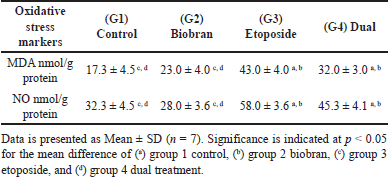 | Table 3. Evaluation of oxidative stress markers (MDA and NO) in rat liver homogenates among experimental groups. [Click here to view] |
Quantification of antioxidants SOD, CAT, and GSH in rat liver cells homogenate from different experimental groups
To investigate the antioxidant activity, SOD, CAT, and GSH peroxidase were measured in rat liver cells homogenate as demonstrated in Table 4. The biobran group did not show any significant differences compared to the control group for any of the three enzymes. In contrast, SOD, CAT, and GSH showed a significant decrease (p < 0.05) in the etoposide group compared to the control group and the biobran group. The dual treatment group showed a substantial reduction in comparison to the control group for CAT, GSH, and SOD. The results indicated a significant increase (p < 0.05) for SOD and GSH but not CAT parameters in the dual treatment compared to the etoposide group.
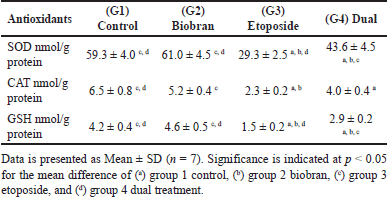 | Table 4. Comparative analysis of antioxidant markers (SOD, CAT, and GSH peroxidase) in rat liver homogenates. Groups. [Click here to view] |
Inflammatory status of the liver: comprehensive evaluation of cytokine profiles
An ELISA assay was used to assess the liver’s inflammatory status by measuring interleukins (IL-6, IL-17, IL-10, IL-4, and IL-8) in rat liver (Table 5). The results clearly demonstrated that biobran treatment alone had no significant effect on the inflammatory profile as compared to the control group, whereas the dual treatment produced varying results.
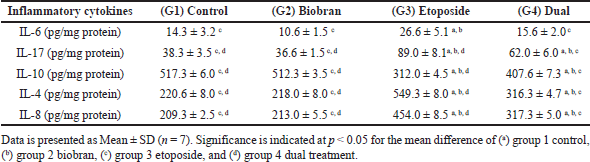 | Table 5. Comparative analysis between experimental groups of pro-inflammatory cytokines (IL-6, IL-17), anti-inflammatory cytokine (IL-10), a key immune regulatory mediator (IL-4), and a chemoattractant mediator (IL-8) in rat liver using ELISA assay. [Click here to view] |
Etoposide treatment significantly increased the proinflammatory cytokine IL-6 (p < 0.05) compared to the control and biobran-treated groups. While, the dual treatment showed a modest decrease in IL-6 levels compared to the etoposide group, but no significant difference was found for dual treatment when compared to the control or biobran groups.
IL-17 levels increased significantly (p < 0.05) after etoposide treatment compared to the control, biobran, and dual-treated groups. Interestingly, the dual treatments showed a significant decrease in IL-17 compared to etoposide. However, IL-17 was still significantly higher (p < 0.05) in dual treatment compared to control and biobran groups.
In contrast, etoposide treatment significantly reduced the anti-inflammatory cytokine IL-10 (p < 0.05) compared to all other groups.
IL-4 and IL-8 levels significantly increased (p < 0.05) in the etoposide group compared to the control, biobran, and dual-treatment groups. Notably, the dual treatment had significantly increased IL-4 and IL-8 levels (p < 0.05) than the control and biobran groups.
Histological and histopathological observations of liver tissue: a comparison of treatments with etoposide, biobran, and dual treatment with both, compared with normal liver
Photomicrographs from both control G1 (Fig. 1A) and biobran G2 (Fig. 1B) depicted normal hepatic architecture, which consists of cords of HC with acidophilic cytoplasm and centrally located round basophilic nuclei (N) radiating outward from a central vein (CV) to the periphery of the lobule. These cords of HC were separated by blood sinusoids (BS) (Fig. 1A and B). Liver sections from rats treated with etoposide (G3) (Fig. 1C–1E) revealed hepatocellular injury in specific areas, evidenced by disorganized hepatic cellular structure and loss of the normal tissue architecture. Some HC exhibited ballooning degeneration and displayed marked vacuolated cytoplasm and pyknotic N, along with dilated and congested veins (Fig. 1C and D). Diffuse and periportal inflammatory leukocytic infiltrations were observed as indicated by an asterisk, along with proliferating bile ducts in Figure 1E. Additionally, activated Kupffer cells were present. Liver sections from the dual treatment (G4) showed a noticeable improvement in HC and liver architecture (Fig. 1F). Histopathological examination indicated mitigation of hepatocellular injury, and a restoration of a more normal liver architecture compared to rats treated with etoposide alone. These histological findings were further supported by the semiquantitative scoring of liver injury parameters presented in Figure 2, which complements the findings from the histopathology in Figure 1. The etoposide group (G3) had increased scores for all the parameters evaluated, including HC degeneration, veinous congestion, necrosis, inflammation, and bile duct proliferation, which confirmed the extensive liver damage observed microscopically. In contrast, the G4 dual treatment group showed significantly reduced pathology scores, consistent with the histological improvement seen in Figure 1F. The biobran-treated group (G2) had scores that were essentially indistinguishable from the control group (G1), reflecting preserved hepatic structure. These results collectively demonstrate the hepatotoxic effect of etoposide and the protective potential of biobran when given.
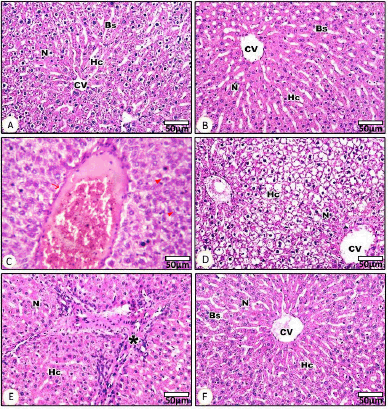 | Figure 1. Histopathological examination of liver tissues stained with hematoxylin and eosin (H&E) under light microscopy (400× magnification). (A–F) Representative photomicrographs showing liver architecture across different experimental groups. (A) Control group (G1): Normal hepatic architecture with radiating cords of polygonal HC around a CV, normal N, and regular BS. (B) Biobran-treated group (G2): Well-preserved liver structure similar to control, with normal HC, CV, and intact BS. (C–E) Etoposide-treated group (G3): Severe histopathological alterations observed. (C) Congested and dilated portal vein. (D) HC vacuolation and cytoplasmic degeneration with loss of normal architecture. (E) Diffuse inflammatory cell infiltration around the portal area (asterisk), HC swelling, and disruption of lobular organization. (F) Dual treatment group (G4): Improved hepatic architecture compared to the etoposide group, showing restoration of HC structure, normal N, and CV, with reduced inflammation and congestion. Scale bar = 50 μm. [Click here to view] |
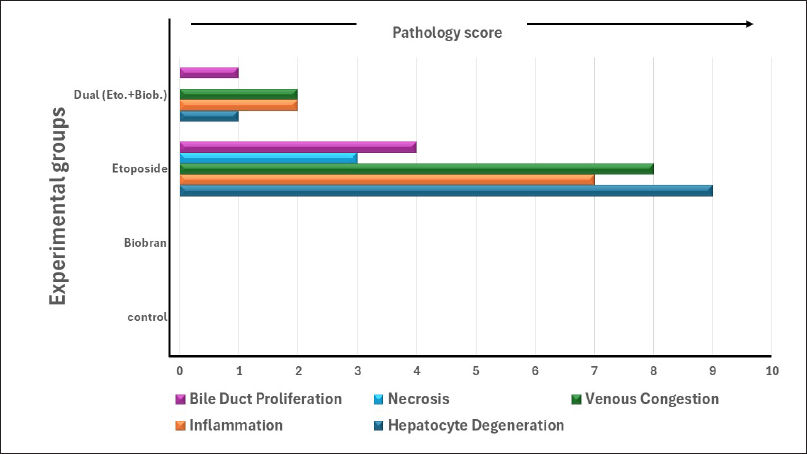 | Figure 2. Semiquantitative histopathological grading of liver tissue changes among experimental groups. Liver sections were evaluated for five main parameters: HC degeneration, inflammation, venous congestion, necrosis, and bile duct proliferation. Each parameter was scored on a 0–3 scale based on the severity (0 = none, 1 = mild, 2 = moderate, 3 = severe). The sum of 4 animals per group is shown. The etoposide group showed the highest pathology scores in all the parameters, indicating severe hepatic injury. Combined treatment with Biobran reduced the pathological changes, while the Biobran-alone group showed normal structure-like control. [Click here to view] |
Immunohistochemical analysis of proinflammatory cytokines IL-1β, TNF-α, and TGF-β in HC
Figure 3 presents light micrographs that demonstrate the effect of biobran treatment on the expression levels of proinflammatory cytokines IL-1β, TNF-α, and TGF-β after etoposide treatment in rat liver tissues. For the control group (Fig. 2 A1–A3), few HC exhibited positive staining for IL-1β (A1), TNF-α (A2), and TGFβ (A3), indicating low basal expression in healthy liver. Similar to the control group, biobran treatment alone (Fig. 2 B1–B3) resulted in minimal staining for all three cytokines, suggesting no difference compared to control. The etoposide group (Fig. 2 C1–C3) showed obvious increased staining intensity for IL-1β (C1), TNFα (C2), and TGF-β (C3) compared to the control and biobran groups (indicated by arrows). Rats treated with both etoposide and biobran (Fig. 2 D1–D3) show a clear decrease in the staining intensity for IL-1β (D1), TNF-α (D2), and TGF-β (D3) compared to the etoposide group.
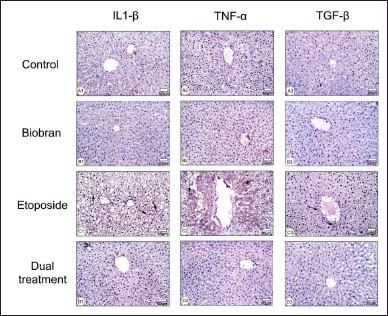 | Figure 3. Micrographs showing the effects of biobran treatment on rat liver tissues following etoposide-induced hepatotoxicity, showing IL-1β, TNF-α, and TGF-β expression in HC. Rats in the control group showed a few HC that were positively stained for IL-1β (A1), TNF-α (A2), and TGF-β (A3) expression. Biobran-treated rats also demonstrated that IL-1β (B1), TNF-α (B2), and TGF-β (B3) are expressed in a few HC. The etoposide-treated rats showed an increase of IL-1β (C1), TNF-α (C2), and TGF-β (C3) compared with the control group. Rats treated with both etoposide and biobran showed HC with a decrease in IL-1β (D1), TNF-α (D2), and TGFβ (D3) relative to the etoposide group. Magnification: 200×. [Click here to view] |
Flow cytometric assessment of apoptotic markers: analysis of caspase-3, Bcl-2, and annexin activities following treatments with etoposide, biobran, and both in liver cells
Flow cytometry analysis was conducted to assess the impact of different treatments in apoptosis including caspase3, Bcl2 annexin-V/PI activities in liver cells (Fig. 4). The flow cytometry analyses were performed four times, and the data reported here reflect an average percentage of the four outcomes.
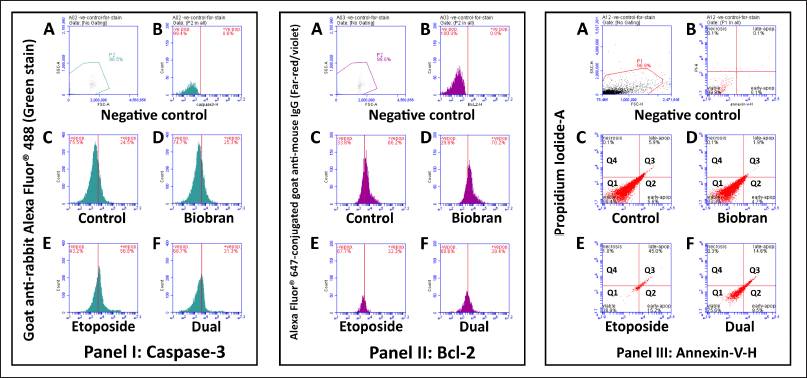 | Figure 4. Flow cytometry analysis of apoptotic markers in liver tissues from experimental groups. Panel I: Caspase-3 expression was assessed using a rabbit anti-Caspase-3 primary antibody followed by goat anti-rabbit Alexa Fluor® 488 secondary antibody (green fluorescence). Panel II: Bcl-2 expression was evaluated using a mouse anti-Bcl-2 primary antibody followed by goat anti-mouse Alexa Fluor® 647 secondary antibody (far-red fluorescence). Panel III: Apoptotic and necrotic cell populations were identified using Annexin V- and PI staining. (A–B) show negative controls and isotype controls used to establish gating boundaries and eliminate background signal. (C–F) represent samples from different experimental groups: (C): Control group. (D): Biobran-treated group. (E): Etoposide-treated group. (F): Dual treatment group (Etoposide + Biobran). Histograms in Panels I and II display the percentage of Caspase-3+ and Bcl-2+ cells. Dot plots in Panel III show quadrants representing viable cells (Annexin-/PI-), early apoptotic (Annexin+/PI-), late apoptotic (Annexin+/PI+), and necrotic cells (Annexin-/PI+). Notably, etoposide-treated livers showed an increased percentage of apoptotic cells (Caspase-3+, Annexin V+) and reduced Bcl-2 expression, while dual treatment mitigated these changes. [Click here to view] |
Caspase-3 activity
Figure 4 panel I shows caspase-3 data as histograms, with the x-axis showing the level of caspase-3 activity and the y-axis showing the number of cells at each level. The histograms are also overlaid with vertical lines that show the gates that were used to define the positive and negative populations. Figure 4A and B shows negative and isotype control. Results from the control group (Fig. 4 Panel I, C) illustrate baseline caspase-3 activity. The percentage of cells in the positive gate (i.e., the cells with caspase-3 activity) is 24.5%, while the caspase-3 negative population is 75.5%, indicative of the normal physiological state of liver cells. In (Fig. 4 Panel I, D) representing the biobran group, caspase-3 positive population was 25.3%. For the etoposide group (Fig. 4 Panel I, E), the percentage of cells in the positive gate is 56.6%, showing an increase compared to the control group. This elevation in caspase-3 activity suggests the induction of apoptosis in response to etoposide treatment, compared to the control and biobran-only treated group. Furthermore, liver tissue from rats treated with both compounds (Fig. 4 Panel I, F) demonstrated a decrease in caspase-3 activity (31%) compared to the etoposide-treated group. This reduction in caspase-3 activity indicates a potential protective effect against etoposide-induced apoptosis when the compounds are both administered.
Bcl2 activity
Figure 4 panel II shows a flow cytometry analysis of Bcl2 activity in rat liver cells. Bcl2 is a protein that helps to protect cells from apoptosis or programmed cell death. Figure 4, panels II A and B illustrate the negative and isotype control. The experimental control group shows the basal level of Bcl2 activity in untreated rat liver with a positive population of 66.2% (Fig. 4 Panel II C). The biobran group, Figure 4 panel II D shows approximately the same BcL2 (70.2%) activity very close to the control. Etoposide group BcL2 activity diminished to (32.3%) as in Figure 4, panel II E, while the dual treatment group (Fig. 4, panel II F) shows a notable increase in Bcl2 activity (39.4%) when compared to the etoposide only group.
Annexin activity
Figure 4, panel III shows scatter plots of annexin activity, representing the negative and isotype control (A and B) and four different experimental treatment groups (C, D, E, and F). Each plot has an axis “Annexin V-PE” for the measured binding of Annexin V dye to the cells (higher Annexin V-PE signal indicates a higher proportion of cells in early apoptosis), and an axis “PI” for the measured uptake of PI dye (higher PI signal indicates a higher proportion of cells in late apoptosis or necrosis). The lower left quadrant Q1 (Annexin V-/PI-) measures viable live cells with intact membranes and DNA. The lower right quadrant Q2 (Annexin V+/PI-) signifies the early stage of apoptosis, where phosphatidylserine is exposed but DNA remains intact. The upper right quadrant Q3 (Annexin V+/PI+) encompasses late apoptosis (both membrane changes and DNA damage). The upper left quadrant Q4 (Annexin V-/PI+) represents cells with compromised membranes but without PS exposure. Its presence could indicate unhealthy cells with early membrane damage. Figure 4, panel III A and B, shows negative control. For the experimental control group (Fig. 4, panel III C), most cells (88.4%) are in Q1, indicating they are viable. A small percentage of cells are in Q2 (5.6%) for early apoptosis, Q3 (5.9%) for late apoptosis, and Q4 (0.1%) for necrosis. Likewise, for the biobran group (Fig. 4, panel III D), most cells (93.3%) are in Q1, indicating viability, the proportion of cells in Q2 (4.7%) and in the proportion of cells in Q3 (1.9%), while Q4 indicating necrosis was only 0.1%. As seen in Figure 4, panel II E, the etoposide group had a decrease in viable cells in Q1 (38.8%) and an increase in Q2 (15.2%) and Q3 (45.0%) cells, indicating a higher proportion of cells in early and late apoptosis. The proportion in Q4 indicating necrosis was 1.0%, suggesting that etoposide treatment induces cell death in this cell population. The panel illustrating measurements for the dual treatment group as in Figure 4, panel III F showed that the majority of cells (75.5%) are in Q1, indicating viability. Compared to etoposide treatment, there is a decline in the proportion of cells in Q2 (9.5%) and in Q3 (14.6%), while Q4 indicating necrosis showed 0.3%. These results suggest that dual treatment with biobran had a protective effect against apoptosis.
Flow cytometric analysis of CD4, CD8, and CD3 molecule expression in rat liver under different treatment conditions including etoposide, biobran, and both
Flow cytometry was employed to examine liver-resident or infiltrating T cell populations according to measurement of CD4+, CD8+, and CD3+ surface antigens in liver single-cell suspensions of rats treated with various regimens: control (G1), biobran (G2), etoposide (G3), and combined biobran plus etoposide treatment (G4). FSC versus SSC gating was initially used to gate the viable single cells and exclude cell clumps and debris. Percentage figures for CD4+, CD8+, and CD3+ cells were subsequently represented as a percentage of the SSC/FSC-gated viable single-cell population. Four rats (n = 4) were in each group, and statistical comparisons were accordingly made (Fig. 5II).
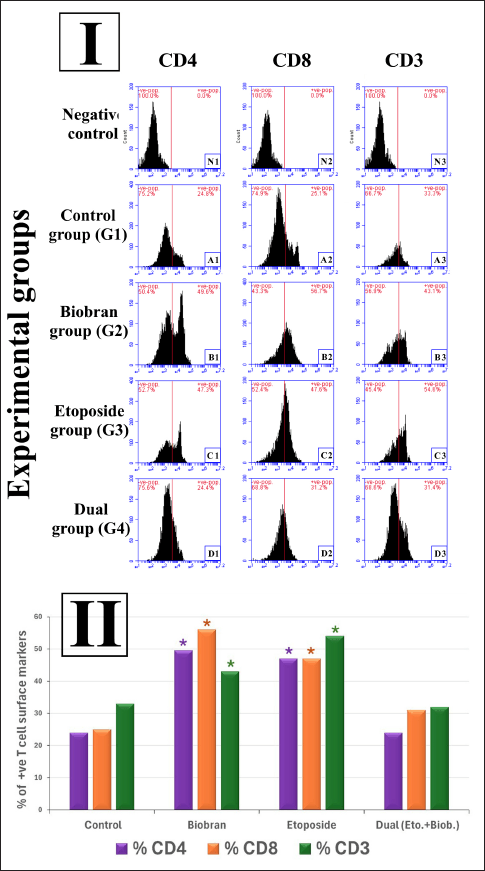 | Figure 5. Flow cytometric analysis of T cell surface markers (CD4, CD8, and CD3) in liver tissues of different experimental groups. (I) Representative histograms showing the expression of CD4, CD8, and CD3 surface markers in liver-derived single-cell suspensions from the following groups: negative control (unstained cells; N1–N3), control group (G1; A1–A3), biobran-treated group (G2; B1–B3), etoposide-treated group (G3; C1–C3), and dual treatment group (G4; D1–D3). Percentages of positive (+ve) and negative (−ve) cell populations are shown in red above each histogram. Staining was performed using fluorochrome-conjugated antibodies, and analysis was conducted using a BD Accuri™ C6 flow cytometer. (II) Bar graph summarizing the mean percentages of positively stained T cells for each marker across experimental groups. A significant increase in CD4+, CD8+, and CD3+ T cell populations was observed in the biobran and etoposide groups compared to control. Dual treatment with biobran and etoposide reduced these populations to near-control levels. Data are presented as means (n = 4); p < 0.05 compared to control (denoted by * above bars). [Click here to view] |
Representative histograms of CD4, CD8, and CD3 staining are shown in Figure 5I, and quantitative data corresponding to them are shown in Figure 5II. N1, N2, and N3 are the respective CD4, CD8, and CD3 gates for the negative controls.
In the control group (G1), the baseline T cell proportions were: CD4+ 24.8%, CD8+ 25.1%, and CD3+ 33.3%. These were utilized as controls to measure the effects of treatment. Administration of biobran alone (G2) resulted in highly significant elevation in proportions of all the T cell subsets with CD4+ reaching 49.6%, CD8+ at 56.7%, and CD3+ at 43.1%, which showed a significant immunostimulatory effect (p < 0.05 vs. control) as in Figure 5II. Etoposide (G3) treatment also led to higher CD4+ and CD8+ T cell levels, but these increased to 47.3% and 47.6%, respectively, and CD3+ increased to 54.6%, which also was significantly increased (p < 0.05 vs. control) in Figure 5II. The combined drug group (G4) also shared similar or lower T cell counts than the control group: CD4+ 24.4%, CD8+ 31.2%, and CD3+ 31.4%, suggesting that co-administration of etoposide and biobran would down-regulate the single immunostimulatory effects exhibited with each drug individually.
DISCUSSION
While chemotherapy remains a mainstay in cancer treatment, its efficacy is often hampered by adverse effects like liver damage and immune modulation. Etoposide, a common chemotherapeutic agent, exemplifies this challenge, presenting significant hepatotoxicity and compromising immune function. Biobran, a natural hemicellulose extract from rice bran, offers a promising strategy for mitigating etoposide-induced liver damage and immune suppression through its potent immunostimulatory effects [31]. Our earlier study examined the chemo-preventive role of biobran on liver carcinogenesis in rats and showed that biobran inhibits hepatocarcinogenesis by mechanisms that include inhibition of inflammation, and suppression of cancer cell proliferation [32]. Investigating the potential synergy between etoposide and biobran provides a great opportunity to improve patient outcomes and reduce side effects associated with cancer treatment.
Initially, we evaluated liver function by measuring AST, ALT, total bilirubin, and albumin in serum. The assay results reveal that etoposide induces significant hepatocellular injury in rats, as evidenced by elevated AST and ALT levels, increased total bilirubin, and decreased albumin. These findings align with previous studies on etoposide-induced hepatotoxicity [33,34]. In contrast, the group treated with both etoposide and biobran (dual treatment group) showed no significant changes in AST and ALT compared to the control, suggesting the potential protective effects of biobran against etoposide-induced liver damage. This protective effect may be attributed to the antioxidants in the dual treatment and the ability of biobran to mitigate oxidative stress, maintain bile flow, and preserve protein synthesis [35].
The assay results also indicated that etoposide markedly increased total bilirubin production, reflecting impaired bile flow or conjugation, while biobran treatment effectively mitigated these alterations. This highlights the potential protective role of biobran against etoposide-induced changes in bilirubin levels, offering a nuanced understanding of their differential impact on specific biochemical markers [36]. In addition, our results revealed that etoposide administration resulted in reduced albumin production, indicating potential liver dysfunction. However, biobran intervention effectively prevented such decreases, suggesting its protective role in maintaining normal albumin levels. These agree with previous research, where the administration of biobran led to a notable reduction in liver preneoplastic lesions, resulting in a restoration of normal hepatocellular structure. This effect was accompanied by the suppression of collagen fiber accumulation [37].
Several investigations have detailed the deleterious effects of chemotherapeutic agents on immune constituents, encompassing T and B cells, and components of innate immunity [38]. This is evident in the diminished levels of IgM and IgG immunoglobulins following etoposide treatment, whereas these levels were restored to normal with biobran. Elsaid et al. [39] illustrated that biobran serves as a potent biological response modifier, stimulating diverse immune system elements such as NK, T, and B cells. Our results corroborate this, confirming biobran’s capacity to modulate immune responses. Etoposide, an antineoplastic agent, exerts its anticancer effects by impeding DNA replication, interacting with topoisomerase, and inducing oxidative stress in tumor cells. Our results similarly demonstrate that etoposide induces oxidative stress, reflected by our measurements of heightened MDA and NO levels as previously mentioned [40,41]. Crucially, the dual treatment and interventions with biobran did not exacerbate oxidative stress, implying potential protection against etoposide-induced oxidative damage. Previous evidence indicates that biobran has the capacity to elevate oxidative stress in the liver while concurrently inhibiting biomarkers such as MDA, total free radicals, and NO levels in murine Ehrlich carcinoma. Biobran induces oncostatic activity by regulating lipid peroxidation, enhancing the antioxidant defense system, and safeguarding against oxidative stress [42].
In the current study, etoposide treatment significantly decreased the activity of all three antioxidant enzymes (SOD, CAT, and GSH) compared to the control group. This indicates that etoposide induces oxidative stress in the rat liver, leading to increased free radical production and depletion of antioxidant defenses [43]. While still significantly lower than the control group, the dual treatment with etoposide and biobran showed a slight increase in SOD and CAT activity compared to the etoposide-only group. This suggests that biobran may offer some protection against etoposide-induced oxidative stress, but further research is needed to confirm its efficacy and underlying mechanisms. Interestingly, biobran supplementation alone maintained SOD, CAT, and GSH at levels similar to the control group. This highlights the potential antioxidant and protective properties of biobran, possibly due to its fiber content and bioactive compounds. The results of this research are largely consistent with and confirm the results of previous research [44].
Regarding liver inflammation, etoposide caused inflammation in the liver, but biobran helped to counteract the inflammatory status, as indicated by measurements of both proinflammatory and anti-inflammatory cytokines by ELISA in the liver. The amounts of proinflammatory cytokines (IL-6 and IL-17) increased after treatment with etoposide, while decreased after using biobran together with etoposide in the dual treatments. A similar but oppositely oriented trend was clear in the case of the anti-inflammatory cytokine, IL-10. Both sets of results confirm the ability of biobran to mitigate the inflammatory actions resulting from etoposide. From our results and previous studies [45], we conclude that combined treatment with some natural ingredients such as biobran could serve as a complementary therapeutic agent in reducing the toxic and inflammatory lesions caused by etoposide. In addition, IL-8, a chemokine that primarily recruits neutrophils to sites of inflammation, increased in the liver from etoposide-treated rats indicating acute inflammation, a case of drug-induced liver injury. IL-8 elevation also indicates a strong neutrophilic response, which can lead to further liver tissue damage [46,47]. IL-4 is a key cytokine in promoting Th2 immune responses and is associated with fibrosis, allergy, and wound healing. An increase in IL-4 in the liver of etoposide treatment indicates a shift toward a Th2-dominated immune response, which can contribute to liver fibrosis through the activation of hepatic stellate cells [48].
The ELISA results measuring inflammatory cytokines in the liver were confirmed by immunohistochemistry results that showed an increase in the expression of proinflammatory cytokines (IL-1β, TNF-α, and TGF-β) in the etoposide-treated group, indicative of hepatotoxicity. Co-treatment with biobran resulted in a decrease in the expression of these cytokines, suggesting a potential protective effect against etoposide-induced inflammation and liver damage. These results align with previous studies that have shown an increase in peripheral levels of TNF-α, IL-1β, and IL-6 following treatment with chemotherapeutic agents, doxorubicin, or cisplatin [49,50].
Moreover, histopathological examinations provide valuable insights into the structural changes in liver tissues. Rats treated with etoposide displayed significant hepatocellular injury, inflammation, and architectural disruption. However, the co-administration of biobran with etoposide demonstrated a protective effect, leading to an observable improvement in HC and liver structure. These findings suggest the potential hepatoprotective properties of biobran in the context of etoposide-induced liver damage. In an earlier study, histopathological alterations were noted following etoposide treatment, including parenchymal ruptures and HC vacuolation. Dilation of BS and congestion of amorphous material in central blood vessels were also observed [51,52].
Regarding apoptosis inducement related to the etoposide mechanism of action, the high percentage of late apoptotic/necrotic cells after etoposide treatment indicated by flow cytometry measurements of caspase 3, annexin, and Bcl2 activities aligns with its known mechanism of action. Etoposide inhibits topoisomerase II, leading to DNA damage and triggering apoptosis via p53 and other pathways. This strong pro-apoptotic effect is consistent with its use in cancer therapy. The higher level of early apoptosis in the dual treatment group compared to the control and biobran groups is intriguing. It suggests that biobran might initially enhance etoposide’s apoptotic cascade. However, the lack of progression to late apoptosis/necrosis indicates that biobran might also interfere with downstream apoptotic events or promote cell survival mechanisms. The similar apoptotic profiles of the control and biobran groups suggest that biobran alone does not significantly trigger or inhibit apoptosis in liver cells. This strengthens the hypothesis that biobran’s potential effect on apoptosis might be limited to modulation when combined with etoposide [18,53].
This study also investigated the impact of etoposide and biobran on T cell infiltration in the liver. Flow cytometry was used to quantify CD4+ (helper T cells) and CD8+ (cytotoxic T cells) populations, alongside CD3+ (general T cell marker) expression. The results show significant elevation of CD4+, CD8+, and CD3+ cells after etoposide treatment in the liver compared to control. This suggests etoposide triggers liver inflammation and oxidative stress, inducing cytokine release and attracting T cells for immune response. Activation of Kupffer cells (liver macrophages) could also contribute to T cell recruitment. Distinctive elevation of CD4+, CD8+, and CD3+ cells even in the absence of etoposide implies that biobran might directly stimulate T cell proliferation and infiltration in the liver, potentially enhancing immune function. Biobran’s immunomodulatory and antioxidant properties might be mediated by multiple probable molecular pathways. One such important approach is the stimulation of the Nrf2 (nuclear factor erythroid 2-related factor 2) pathway, which induces the antioxidant response and increases the expression of endogenous antioxidant enzymes such as SOD, CAT, and GSH. Biobran may also inhibit the NF-κB pathway and reduce the expression of pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-6, hence supporting its role as a natural immunomodulator and antioxidant. Further research is required for understanding the specific mechanisms of biobran’s T cell activation and its impact on immune effectiveness [54].
Our findings might be explained by considering that DNA damage from etoposide treatment may activate stress pathways in liver cells, triggering inflammatory responses and cytokine production. These cytokines, like IL-1 and TNF-α, can attract and activate T cells to infiltrate the liver and participate in tissue repair or damage control. Also, Kupffer cells, recognizing the stress signals, can directly activate T cells through antigen presentation and cytokine release. The ability of biobran to enhance hepatic T cell infiltration can be attributed to its systemic immunomodulatory actions. Biobran has been shown to activate innate immune responses and cause maturation of dendritic cells and cytokine release, including interleukins and interferons, that can promote the recruitment and activation of T cells [55]. In addition, immunomodulators administered intraperitoneally are demonstrated to stimulate peritoneal immune cells, including macrophages and dendritic cells, which are capable of migrating to the liver and triggering hepatic immune response [56]. Thus, the increased T cell infiltration in the liver may reflect the direct systemic immunostimulatory effect of Biobran. Biobran’s antioxidant properties might also protect T cells from oxidative damage, promoting their survival and activity [57,58].
CONCLUSION
We investigated the combined effects of biobran (a dietary fiber supplement) and etoposide (a common chemotherapeutic agent) on liver health and immune function. Our findings suggest that biobran holds substantial promise as a complementary therapy to protect against etoposide-induced liver damage and immune suppression. This potentially offers patients a safer and more effective path to overcome both the disease and its adverse side effects. The exploration of the synergy between etoposide and biobran represents a notable opportunity to enhance patient outcomes and minimize the complications associated with cancer treatment.
ACKNOWLEDGMENTS
The authors would like to take this opportunity to express their warm appreciation to the members of the Animal House, Faculty of Science, Damanhour University, for their useful aid and support in experimental animal handling and care during the duration of this research.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
This research did not receive any specific grant from funding agencies in the public, commercial, or non-profit sectors.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Ethical approvals details are given in the ‘Materials and Methods’ section.
CONSENT FOR PUBLICATION
All authors have evaluated the work and approved its publication in this journal. Each author acknowledges approving the final version and is accountable for all elements of the study, including ensuring that questions of accuracy and integrity are properly addressed.
DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Lee EM, Jiménez-Fonseca P, Galán-Moral R, Coca-Membribes S, Fernández-Montes A, Sorribes E, et al. Toxicities and quality of life during cancer treatment in advanced solid tumors. Curr Oncol. 2023;30(10):9205–16. CrossRef
2. Anand U, Dey A, Chandel AKS, Sanyal R, Mishra A, Pandey DK, et al. Cancer chemotherapy and beyond: current status, drug candidates, associated risks and progress in targeted therapeutics. Genes Dis. 2023;10(4):1367–401. CrossRef
3. Money ME, Matthews CM, Tan-Shalaby J. Review of under-recognized adjunctive therapies for cancer. Cancers (Basel). 2022;14(19):4780. CrossRef
4. Eskandari S, Barzegar A, Mahnam K. Absorption of daunorubicin and etoposide drugs by hydroxylated and carboxylated carbon nanotube for drug delivery: theoretical and experimental studies. J Biomol Struct Dyn. 2022;40(20):10057–64. CrossRef
5. Fan YW, Liu MH, Xu TJ, Fan RY, Xiang J, Wu JQ, et al. Mechanism of etoposide resistance in small cell lung cancer and the potential therapeutic options. Med Oncol. 2025;42(5):167. CrossRef
6. Reyhanoglu G, Tadi P. Etoposide. Treasure Island, FL: StatPearls Publishing; 2025.
7. Montecucco A, Zanetta F, Biamonti G. Molecular mechanisms of etoposide. EXCLI J. 2015;14:95–108.
8. Zhang W, Gou P, Dupret JM, Chomienne C, Rodrigues-Lima F. Etoposide, an anticancer drug involved in therapy-related secondary leukemia: enzymes at play. Transl Oncol. 2021;14(10):101169. CrossRef
9. Morsi RM, Mansour DS, Mousa AM. Ameliorative potential role of Rosmarinus officinalis extract on toxicity induced by etoposide in male albino rats. Braz J Biol. 2022;84:e258234. CrossRef
10. Tran A, Housset C, Boboc B, Tourani JM, Carnot F, Berthelot P. Etoposide (VP 16-213) induced hepatitis. Report of three cases following standard-dose treatments. J Hepatol. 1991;12(1):36–9. CrossRef
11. Bailly C. Etoposide: a rider on the cytokine storm. Cytokine. 2023;168:156234. CrossRef
12. Ooi SL, McMullen D, Golombick T, Nut D, Pak SC. Evidence-based review of BioBran/MGN-3 arabinoxylan compound as a complementary therapy for conventional cancer treatment. Integr Cancer Ther. 2018;17(2):165–78. CrossRef
13. Agrawal S, Agrawal A, Ghoneum M. Biobran/MGN-3, an arabinoxylan rice bran, exerts anti-COVID-19 effects and boosts immunity in human subjects. Nutrients. 2024;16(6):881. CrossRef
14. Spaggiari M, Dall’Asta C, Galaverna G, Del Castillo Bilbao MD. Rice bran by-product: from valorization strategies to nutritional perspectives. Foods. 2021;10(1):85. CrossRef
15. Badr El-Din NK, Abdel Fattah SM, Pan D, Tolentino L, Ghoneum M. Chemopreventive activity of MGN-3/Biobran against chemical induction of glandular stomach carcinogenesis in rats and its apoptotic effect in gastric cancer cells. Integr Cancer Ther. 2016;15(4):Np26–34. CrossRef
16. Ashrafujjaman M, Mahtab MA, Noor-E-Alam SM, Rahim MA, Das DC, Ahmed F, et al. Role of biobran (Arabinoxylan Rice Bran) on patients with advanced stage hepatocellular carcinoma. Euroasian J Hepatogastroenterol. 2023;13(2):84–8. CrossRef
17. Bryan LJ, Casulo C, Allen PB, Smith SE, Savas H, Dillehay GL, et al. Pembrolizumab added to ifosfamide, carboplatin, and etoposide chemotherapy for relapsed or refractory classic Hodgkin lymphoma: a multi-institutional phase 2 investigator-initiated nonrandomized clinical trial. JAMA Oncol. 2023;9(5):683–91. CrossRef
18. Badr El-Din NK, Areida SK, Ahmed KO, Ghoneum M. Arabinoxylan rice bran (MGN-3/Biobran) enhances radiotherapy in animals bearing Ehrlich ascites carcinoma†. J Radiat Res. 2019;60(6):747–58. CrossRef
19. Moradi M, Hashemian MA, Faramarzi A, Goodarzi N, Hashemian AH, Cheraghi H, et al. Therapeutic effect of sodium alginate on bleomycin, etoposide and cisplatin (BEP)-induced reproductive toxicity by inhibiting nitro-oxidative stress, inflammation and apoptosis. Sci Rep. 2024;14(1):1565. CrossRef
20. Gurina TS, Simms L. Histology, staining. Treasure Island, FL: StatPearls Publishing; 2025
21. Leow WQ, Chan AW, Mendoza PGL, Lo R, Yap K, Kim H. Non-alcoholic fatty liver disease: the pathologist’s perspective. Clin Mol Hepatol. 2023;29(Suppl):S302–18. CrossRef
22. Huang XJ, Choi YK, Im HS, Yarimaga O, Yoon E, Kim HS. Aspartate aminotransferase (AST/GOT) and alanine aminotransferase (ALT/GPT) detection techniques. Sensors (Basel). 2006;6(7):756–82. CrossRef
23. Ueno T, Hirayama S, Ito M, Nishioka E, Fukushima Y, Satoh T, et al. Albumin concentration determined by the modified bromocresol purple method is superior to that by the bromocresol green method for assessing nutritional status in malnourished patients with inflammation. Ann Clin Biochem. 2013;50(Pt 6):576–84. CrossRef
24. Mori L. Modified Jendrassik--Grof method for bilirubins adapted to the Abbott bichromatic analyzer. Clin Chem. 1978;24(10):1841–5. CrossRef
25. Vos JG, Buys J, Beekhof P, Hagenaars AM. Quantification of total IgM and IgG and specific IgM and IgG to a thymus-independent (LPS) and a thymus-dependent (tetanus toxoid) antigen in the rat by enzyme-linked immunosorbent assay (ELISA). Ann N Y Acad Sci. 1979;320:518–34. CrossRef
26. Arsenijevic D, Stojanovic B, Milovanovic J, Arsenijevic A, Simic M, Pergal M, et al. Hepatoprotective effect of mixture of dipropyl polysulfides in concanavalin A-induced hepatitis. Nutrients. 2021;13(3):1022. CrossRef
27. Luan X, Chen P, Li Y, Yuan X, Miao L, Zhang P, et al. TNF-α/IL-1β-licensed hADSCs alleviate cholestatic liver injury and fibrosis in mice via COX-2/PGE2 pathway. Stem Cell Res Ther. 2023;14(1):100. CrossRef
28. de Souza Dantas Oliveira SH, de Souza Aarão TL, da Silva Barbosa L, Souza Lisbôa PG, Tavares Dutra CD, Margalho Sousa L, et al. Immunohistochemical analysis of the expression of TNF-alpha, TGF-beta, and caspase-3 in subcutaneous tissue of patients with HIV lipodystrophy syndrome. Microb Pathog. 2014;67–68:41–7. CrossRef
29. Wlodkowic D, Skommer J, Darzynkiewicz Z. Flow cytometry-based apoptosis detection. Methods Mol Biol. 2009;559:19–32. CrossRef
30. Bernardo I, Mancebo E, Aguiló I, Anel A, Allende LM, Guerra-Vales JM, et al. Phenotypic and functional evaluation of CD3+CD4-CD8- T cells in human CD8 immunodeficiency. Haematologica. 2011;96(8):1195–203. CrossRef
31. Elsaid AF, Shaheen M, Ghoneum M., biobran/MGN-3, an arabinoxylan rice bran, enhances NK cell activity in geriatric subjects: a randomized, double-blind, placebo-controlled clinical trial. Exp Ther Med. 2018;15(3):2313–20. CrossRef
32. Badr El-Din NK, Ali DA, Othman R, French SW, Ghoneum M. Chemopreventive role of arabinoxylan rice bran, MGN-3/Biobran, on liver carcinogenesis in rats. Biomed Pharmacother. 2020;126:110064. CrossRef
33. Mohammadzadeh S, Kiani A, Amiri M. Lycopene modulates hepatic toxicity and testicular injury induced by etoposide in male rats. F S Sci. 2023;4(1):30–35. CrossRef
34. Tousson E, Bayomy MF, Ahmed AA. Rosemary extract modulates fertility potential, DNA fragmentation, injury, KI67 and P53 alterations induced by etoposide in rat testes. Biomed Pharmacother. 2018;98:769–74. CrossRef
35. Ooi SL, Micalos PS, Pak SC. Modified rice bran Arabinoxylan by Lentinus edodes mycelial enzyme as an immunoceutical for health and aging-a comprehensive literature review. Molecules. 2023;28(17):6313. CrossRef
36. Ooi SL, Pak SC, Micalos PS, Schupfer E, Lockley C, Park MH, et al. The health-promoting properties and clinical applications of rice bran Arabinoxylan modified with Shiitake mushroom enzyme-a narrative review. Molecules. 2021;26(9):2539. CrossRef
37. Hsu FY, Yang SC, Suk FM, Shirakawa H, Chiu WC, Liao YJ. Dietary rice bran attenuates hepatic stellate cell activation and liver fibrosis in mice through enhancing antioxidant ability. J Nutr Biochem. 2024;125:109565. CrossRef
38. Li Z, Lin A, Gao Z, Jiang A, Xiong M, Song J, et al. B-cell performance in chemotherapy: unravelling the mystery of B-cell therapeutic potential. Clin Transl Med. 2024;14(7):e1761. CrossRef
39. Elsaid AF, Agrawal S, Agrawal A, Ghoneum M. Dietary supplementation with biobran/MGN-3 increases innate resistance and reduces the incidence of influenza-like illnesses in elderly subjects: a randomized, double-blind, placebo-controlled pilot clinical trial. Nutrients. 2021;13(11):4133. CrossRef
40. Koushki M, Khedri A, Aberomand M, Akbari Baghbani K, Mohammadzadeh G. Synergistic anti-cancer effects of silibinin-etoposide combination against human breast carcinoma MCF-7 and MDA-MB-231 cell lines. Iran J Basic Med Sci. 2021;24(9):1211–9.
41. Cao M, Lu H, Yan S, Pang H, Sun L, Li C, et al. Apatinib plus etoposide in pretreated patients with advanced triple-negative breast cancer: a phase II trial. BMC Cancer. 2023;23(1):463. CrossRef
42. Zhao Z, Cheng W, Qu W, Wang K. Arabinoxylan rice bran (MGN-3/Biobran) alleviates radiation-induced intestinal barrier dysfunction of mice in a mitochondrion-dependent manner. Biomed Pharmacother. 2020;124:109855. CrossRef
43. Vallejo MJ, Salazar L, Grijalva M. Oxidative stress modulation and ROS-mediated toxicity in cancer: a review on in vitro models for plant-derived compounds. Oxid Med Cell Longev. 2017;2017:4586068. CrossRef
44. Noaman E, Badr El-Din NK, Bibars MA, Abou Mossallam AA, Ghoneum M. Antioxidant potential by arabinoxylan rice bran, MGN-3/biobran, represents a mechanism for its oncostatic effect against murine solid Ehrlich carcinoma. Cancer Lett. 2008;268(2):348–59. CrossRef
45. Jiang H, Geng D, Liu H, Li Z, Cao J. Co-delivery of etoposide and curcumin by lipid nanoparticulate drug delivery system for the treatment of gastric tumors. Drug Deliv. 2016;23(9):3665–73. CrossRef
46. Yang M, Zhang CY. Interleukins in liver disease treatment. World J Hepatol. 2024;16(2):140–5. CrossRef
47. Zimmermann HW, Seidler S, Gassler N, Nattermann J, Luedde T, Trautwein C, et al. Interleukin-8 is activated in patients with chronic liver diseases and associated with hepatic macrophage accumulation in human liver fibrosis. PLoS One. 2011;6(6):e21381. CrossRef
48. Briukhovetska D, Dörr J, Endres S, Libby P, Dinarello CA, Kobold S. Interleukins in cancer: from biology to therapy. Nat Rev Cancer. 2021;21(8):481–99. CrossRef
49. Keeney JTR, Ren X, Warrier G, Noel T, Powell DK, Brelsfoard JM, et al. Doxorubicin-induced elevated oxidative stress and neurochemical alterations in brain and cognitive decline: protection by MESNA and insights into mechanisms of chemotherapy-induced cognitive impairment (“chemobrain”). Oncotarget. 2018;9(54):30324–39. CrossRef
50. Wang C, Zhang F, Cao Y, Zhang M, Wang A, Xu M, et al., Etoposide induces apoptosis in activated human hepatic stellate cells via ER stress. Sci Rep. 2016;6:34330. CrossRef
51. Yang SH, Choi HG, Lim SJ, Lee MG, Kim SH. Effects of morin on the pharmacokinetics of etoposide in 7,12-dimethylbenz[a]anthracene-induced mammary tumors in female Sprague-Dawley rats. Oncol Rep. 2013;29(3):1215–23. CrossRef
52. Noormohammadi M, Ghorbani Z, Shahinfar H, Shidfar F. Is there any hepatic impact associated with rice bran arabinoxylan compound supplementation? A systematic review and dose-response meta-analysis of randomized controlled trials. Clin Nutr ESPEN. 2023;57:665–75. CrossRef
53. Cattrini C, Capaia M, Boccardo F, Barboro P. Etoposide and topoisomerase II inhibition for aggressive prostate cancer: data from a translational study. Cancer Treat Res Commun. 2020;25:100221. CrossRef
54. Nakazawa A, Nakano N, Fukuda A, Sakamoto S, Imadome K, Kudo T, et al. Use of serial assessment of disease severity and liver biopsy for indication for liver transplantation in pediatric Epstein-Barr virus-induced fulminant hepatic failure. Liver Transpl. 2015;21(3):362–8. CrossRef
55. Ooi SL, Micalos PS, Kim J, Pak SC. Rice bran arabinoxylan compound as a natural product for cancer treatment - an evidence-based assessment of the effects and mechanisms. Pharm Biol. 2024;62(1):367–93. CrossRef
56. Yu Y, Zhang J, Wang J, Sun B. The anti-cancer activity and potential clinical application of rice bran extracts and fermentation products. RSC Adv. 2019;9(31):18060–9. CrossRef
57. Ghoneum MH, El Sayed NS. Protective effect of biobran/MGN-3 against sporadic alzheimer’s disease mouse model: possible role of oxidative stress and apoptotic pathways. Oxid Med Cell Longev. 2021;2021:8845064. CrossRef
58. Ghoneum M, Agrawal S. Mgn-3/biobran enhances generation of cytotoxic CD8+ T cells via upregulation of dec-205 expression on dendritic cells. Int J Immunopathol Pharmacol. 2014;27(4):523–30. CrossRef