INTRODUCTION
Diabetes is a significant global health challenge as its prevalence steadily rises annually. The global prevalence of type 2 diabetes mellitus (T2DM) in adults is anticipated to increase from 8.8% in 2017 to 9.9% by 2045, with a more rapid increase in low- and middle-income countries than in high-income countries [1,2]. Indonesia, for instance, is expected to see its diabetes prevalence surge from 9.19% in 2020 to 16.09% by 2045 [3]. Most diabetes cases are T2DM (90%–95%), marked by insulin resistance, leading to dyslipidemia, endothelial dysfunction, elevated inflammatory markers, hyperglycemia, hypertension, and atherosclerosis [4,5].
Insulin resistance remains a key factor in the development of T2DM. Numerous research has investigated the involvement of insulin target tissue such as adipose tissue in the etiopathogenesis of T2DM. Adipose tissue is an important insulin target as it regulates lipid and glucose metabolism and homeostasis [6]. Gustafson et al. [7] stated that adipose tissue expansion through hyperplasia can prevent insulin resistance. This occurs because adipose tissue expansion via hyperplasia can prevent adipocyte hypertrophy, which is associated with insulin resistance [7]. In an in vivo study, inhibiting adipose tissue hyperplasia in lipoatrophic or lipodystrophic animal models actually led to significant insulin resistance, ectopic liver fat accumulation, and reduced glucose tolerance, similar to what is observed in human lipoatrophic/lipodystrophic diabetes [8]. Adipose tissue hyperplasia occurs through adipogenesis, resulting in adipocytes containing lipids. Thus, lipid droplets become the dominant feature of the cells [9].
The genes involved in controlling glycemic homeostasis and insulin resistance in adipose tissue are Cebpa and Slc2a4 [gene encoding glucose transporter type 4 (GLUT4) protein]. The overexpression of Cebpa has been known to be able to promote adipogenesis in cultured fibroblasts, activate insulin receptors (IRs), and increase insulin signaling genes such as Aebp1, Adra1d, Cfd (adipsin), Igfbp, Prkcg, Retn (resistin), and Ucp1 in female rats induced with a high-fat diet [10,11]. Conversely, the repression of Cebpa expression is known to inhibit adipogenesis and lower insulin signaling, leading to insulin resistance [12–14]. Cebpa is known to increase the expression of the Slc2a4 gene and GLUT4 protein, a glucose transporter protein responsive to insulin [15]. The downregulation of the GLUT4 expression in adipose tissue is linked to insulin resistance and T2DM [16,17]. Thus, interventions targeting the Cebpa and Slc2a4 gene expression in adipocyte differentiation hold promise for insulin resistance management in T2DM.
Gene expression can also be influenced by epigenetic processes, including DNA methylation, which can affect the expression of methylated genes without altering DNA sequences [18]. DNA methylation often occurs on cytosine within cytosine-guanine dinucleotides (CpG sites). Genomic regions with the highest CpG density, known as CpG islands, are frequently clustered at gene regulatory locations such as promoter regions [19,20].
Various therapeutic approaches have been used for T2DM management, including promoting insulin secretion by the pancreas, improving insulin sensitivity to target receptors, or increasing glucose uptake in adipocytes [21]. This includes the utilization of natural compounds as therapeutic agents for TDM2, and among them is the oyster mushroom (Pleurotus ostreatus), a widely consumed plant worldwide [22]. Oyster mushroom contains low fat, high fiber, and high protein, making it ideal for preventing hyperglycemia in T2DM. It also contains various bioactive compounds including phenolics, flavonoids, alkaloids, tannins, lectins, laccase, vitamin C, ergosterol, and polysaccharides such as β-glucan, making it a plant with a wide range of pharmacological effects, including antidiabetic, hypolipidemic, hypoglycemic, antioxidant, anti-inflammatory, antitumor, antibacterial, nephroprotective, and hepatoprotective activities [23–25].
Choudhury et al. [26] demonstrated that adding 3 g of oyster mushroom powder to food for 3 months can reduce fasting glucose levels and HbAc1 levels in 27 T2DM and hypertension patients. This effect was achieved through several mechanisms, including activating glucokinase, stimulating insulin spikes, and inhibiting glycogen synthase kinase, which increases glycogen synthesis [26]. Another study by Asrafuzzaman et al. [27] suggested that oyster mushroom powder supplementation improves insulin sensitivity by increasing AMP-activated protein kinase phosphorylation and GLUT4 expression in muscle and adipose tissues, enhancing the glucose uptake in diabetic rats [27]. This effect is consistent with the findings of several in vivo studies stating that a Pleurotus ostreatus ethanolic extract and the polysaccharides contained in P. ostreatus have anti-diabetic effects [25,28]. Xiong et al. [29] found that the ergosterol content in P. ostreatus can act as a hypoglycemic agent by modulating the insulin signaling pathway, thus stimulating the translocation and expression of GLUT4 in the L6 cell line [29]. However, the effect of the oyster mushroom ethanolic extract on the genes involved in glycemic control in the adipose tissue is unknown.
This study was conducted using 3T3-L1 preadipocytes, which exhibited Cebpa and Slc2a4 gene expression and promoted the differentiation of preadipocytes into adipocytes after induction by pro-adipogenic factors consisting of methylisobutylxanthine, dexamethasone, and insulin (MDI) [30]. This study aimed to evaluate the effect of the oyster mushroom ethanolic extract on lipid accumulation, mRNA expression of Cebpa and Slc2a4, and methylation level on Slc2a4 promoter during 3T3-L1 adipocyte differentiation, which could provide valuable insights into their therapeutic potential for insulin resistance in T2DM.
MATERIALS AND METHODS
Preparation of oyster mushroom ethanolic extract
The oyster mushrooms were obtained from agricultural sources in Lembang, West Java, Indonesia. They were formally identified by Joko Kusmoro from the Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Padjadjaran, Indonesia (identification number: 465/LBM/IT/XI/2024). In all, 80 g of dried oyster mushroom powder underwent extraction via the maceration method, repeated three times in 800 ml of ethanol for 4 hours each time. The extract was filtered using Advantec No. 5c filter paper (Advantec MFS, USA) and then evaporated. Following evaporation, an oily residue weighing 1.68 g was obtained and diluted in dimethyl sulfoxide (DMSO, Sigma, USA) to create extracts with concentrations of 25, 50, and 100 μg/ml. The DMSO concentration in the solution was limited to 0.5% for all experiments.
Cell culture and oyster mushroom ethanolic extract treatment
3T3-L1 preadipocytes were a gift from Dr. Afiat Berbudi (Department of Biomedical Sciences, Faculty of Medicine, Universitas Padjadjaran, Indonesia), and the usage of this cell line in this study was approved by The Research Ethics Committee of Universitas Padjadjaran (number 1186/UN6.KEP/EC/2024). The culture and differentiation protocol followed the method developed by Ariyanto et al. [31]. Cells were seeded into four different media in 12-well plates with varying treatments: a negative control medium and media containing the oyster mushroom ethanolic extract at concentrations of 25, 50, and 100 μg/ml, respectively. The 3T3-L1 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, USA, cat no. 11995065) with 10% fetal bovine serum (FBS, Sigma, USA) and 1% penicillin–streptomycin at 37°C with 5% CO. Cells were allowed to grow for 48 hours or until reaching 100% confluence.
After attaining confluence, the cells were incubated in DMEM for an additional 48 hours. To induce differentiation, an MDI cocktail containing 0.5-mM methylisobutylxanthine (Sigma, USA, cat no. 15879), 1-µM dexamethasone (Sigma, USA, cat no. D4902), and 10-µg/ml insulin (Sigma, USA, cat no. I0516) was added to the well plate, which was considered day 0 of the experiment. The cells were then cultured in a medium containing MDI and DMEM for 48 hours. Thereafter, the wells’ media were replaced with 10-μg/ml insulin-containing DMEM. The cells were cultured under the same conditions until day 12 of the experiment. The media were replaced every 48 hours throughout the experiment. The oyster mushroom ethanolic extract was added according to the concentration of each well plate from 24 hours after cell seeding until day 12 experiment. On day 12, oil red O (ORO) staining, RNA extraction, and DNA isolation were performed for lipid accumulation measurement, quantitative RT-PCR (qRT-PCR) analysis, and pyrosequencing, respectively.
ORO staining
The 3T3-L1 cells were stained using ORO on day 12 of differentiation, following the method previously described by Ariyanto et al. [31]. In brief, the cells were fixed with 4% formaldehyde in phosphate-buffered saline for 10 minutes and were stained in a freshly prepared ORO solution (0.18% (wt/vol) ORO in 60% isopropanol) for 15–20 minutes; then, the stain was removed. The lipid accumulation was observed both macroscopically and microscopically (using Olympus CK40) with 40× and 100× magnification [31].
qRT-PCR and gene expression analysis
qRT-PCR was used to assess the mRNA expression of Cebpa and Slc2a4. Quick-RNA™ cDNA Synthesis Kit (Bioline Reagents Ltd., UK) was used to extract total RNA from the 3T3-L1 cell line, following the manufacturer’s instructions. Subsequently, qRT-PCR was performed using the SensiFast™ SYBR® No-ROX kit (Bioline Reagents Ltd., UK) as per the manufacturer’s protocol. Polymerase activation was carried out at 95°C for 2 minutes, followed by 40 cycles of denaturation at 95°C for 5 seconds and annealing or extension at 60°C–65°C for 20 seconds. The expression of the Cebpa and Slc2a4 gene was analyzed using the Livak method with the formula 2-ΔΔCt [31]. The Cebpa and Slc2a4 mRNA expression levels were normalized to the housekeeping gene Gapdh. Due to sample constraints, qRT-PCR experiments were conducted in duplicate rather than triplicate. However, each sample was run in parallel under consistent experimental conditions to ensure technical precision. We acknowledge that triplicate measurements are the standard for reproducibility and have noted this as a limitation in the study. Table 1 presents the list of primer sequences used in qRT-PCR.
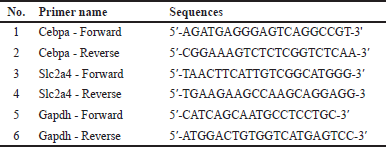 | Table 1. Primer sequence in qRT-PCR. [Click here to view] |
Pyrosequencing analysis
Pyrosequencing was performed to measure the DNA methylation level on the Slc2a4 promoter. DNA isolation from the cells was conducted using the Quick-DNA™ Miniprep Plus kit (Zymo Research, USA), following the manufacturer’s instructions. EZ DNA Methylation-Lightning™ Kit (Zymo Research, USA) was used to perform the bisulfite conversion of DNA, followed by amplification via PCR with one biotinylated primer. PCR amplification utilized the PyroMark PCR kit (Qiagen, Hilden, Germany). Initial PCR activation was set at 95°C for 15 min, followed by 45 cycles of denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, extension at 72°C for 30 seconds, and final extension at 72°C for 10 minutes. Then, 2% agarose gel electrophoresis was used for confirming the PCR reactions and the sizes of the product. Biotinylated PCR products (10 µl) were combined with streptavidin-coated sepharose beads (1 μl) and a PyroMark binding buffer (40 µl) for a total reaction volume of 80 µl. The PCR products were then purified by passing the mixture through PyroMark Q96 Vacuum Workstation (Qiagen). Subsequently, the purified PCR products were combined with an annealing buffer that contained the sequencing primer. Following annealing, the plate was placed in the PyroMark Q96 MD instrument (Qiagen). The proportion of DNA methylation at the CpG sites was quantified using the PyroMark software. The PyroMark assay design program (v.2.0; Qiagen, Germantown, MD, USA) was used for constructing specific primers. The region between −325 and −1 in the proximal Slc2a4 promoter was the target for the methylation level. Percentages of methylation indicate the ratio of signal intensities for C and T at every C within a CpG site. Pyrosequencing was performed in triplicate. Table 2 presents the primers and sequences for analysis by pyrosequencing.
 | Table 2. Primer and sequence to analyze used for pyrosequencing analysis. [Click here to view] |
Statistical analysis
Statistical analysis was performed in SPSS version 26 for Windows (IBM software, New York) and graphs were created using GraphPad Prism version 8.3 for Windows (GraphPad software, Inc., San Diego, CA). The Kruskal–Wallis or One-Way ANOVA tests were used to analyze differences between groups after assessing normality with the Shapiro-Wilk test and homogeneity with the Levene test. Results were presented as mean ± SD. Statistical significance was defined as a p < 0.05.
RESULTS
Oyster mushroom ethanolic extract increases lipid accumulation during adipocyte differentiation
The ORO staining of lipid droplets in the 3T3-L1 cells was used for investigating the effect of oyster mushroom ethanolic extract on lipid accumulation during adipocyte differentiation. Macroscopically, samples treated with the 50- μg/ml oyster mushroom ethanolic extract contained the most lipid droplets, as shown by the red stain. Microscopically, the lipid accumulation observed under 40× and 100× magnification showed that samples treated with the 50-μg/ml ethanolic extract had the highest lipid accumulation. Lipid accumulation at concentrations of 25 μg/ml appeared to be less but still more than that in the case of the control (0 μg/ml) (Fig. 1). This finding indicated the ability of the oyster mushroom ethanolic extract to increase lipid accumulation during adipocyte differentiation.
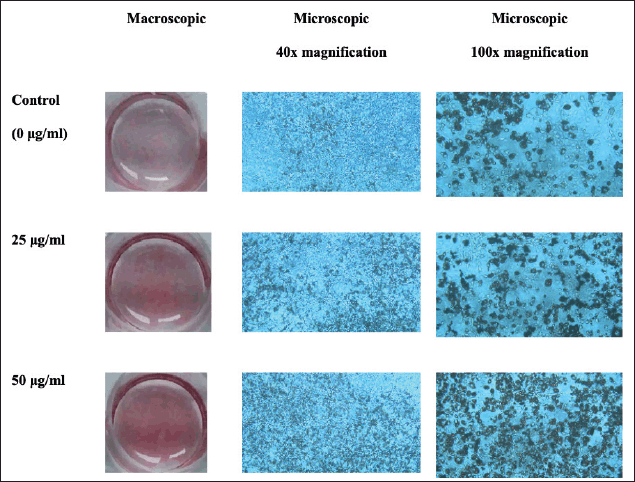 | Figure 1. Macroscopic and microscopic pictures of 3T3-L1 cells after ORO staining on day 12 following treatment with oyster mushroom ethanolic extract. Control (0 µg/ml): cells treated with the same concentration of ethanol without oyster mushroom extract. [Click here to view] |
Oyster mushroom ethanolic extract increased Cebpa and Slc2a4 gene expression
The mRNA expression of Cebpa and Slc2a4 was measured in samples treated with 0, 25, and 50 µg/ml of the oyster mushroom ethanolic extract. The results indicated that the oyster mushroom ethanolic extract increased the expression of the Cebpa and Slc2a4 gene in a dose-dependent manner. Treatment with the oyster mushroom ethanolic extract at the 50-μg/ml concentration showed the highest Cebpa and Slc2a4 mRNA expression levels (about two-fold and fifty-fold increase as compared to those observed in the case of the control, respectively) (Fig. 2). The significant difference in Cebpa and Scl2a4 expression between groups was statistically analyzed using Kruskal–Wallis. The statistical analysis showed no significant difference among all groups (p > 0.05).
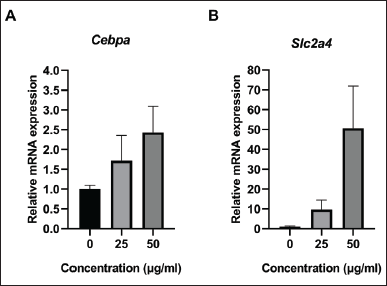 | Figure 2. The relative mRNA expression of (A) Cebpa and (B) Slc2a4 in 3T3-L1 cell line on day 12 following treatment with oyster mushroom ethanolic extract. mRNA expression was measured using qRT-PCR and normalized to Gapdh. The experiment was performed in duplicate. Kruskal-Wallis analysis indicated no significant difference between treatment groups (p > 0.05). Data are presented as mean ± SD. 0 µg/ml (Control): cells treated with the same concentration of ethanol without oyster mushroom extract. [Click here to view] |
Oyster mushroom ethanolic extract decreased methylation level on Slc2a4 promoter
The DNA methylation level on the Slc2a4 promoter was analyzed to investigate the involvement of epigenetic processes, particularly DNA methylation, in regulating the expression of the Slc2a4. Pyrosequencing was performed in samples treated with 0-, 25-, 50-, and 100-μg/ml oyster mushroom ethanolic extracts. Three CpG sites were identified on the Slc2a4 promoter within the designed sequence range: CpG1, CpG2, and CpG3 (Fig. 3).
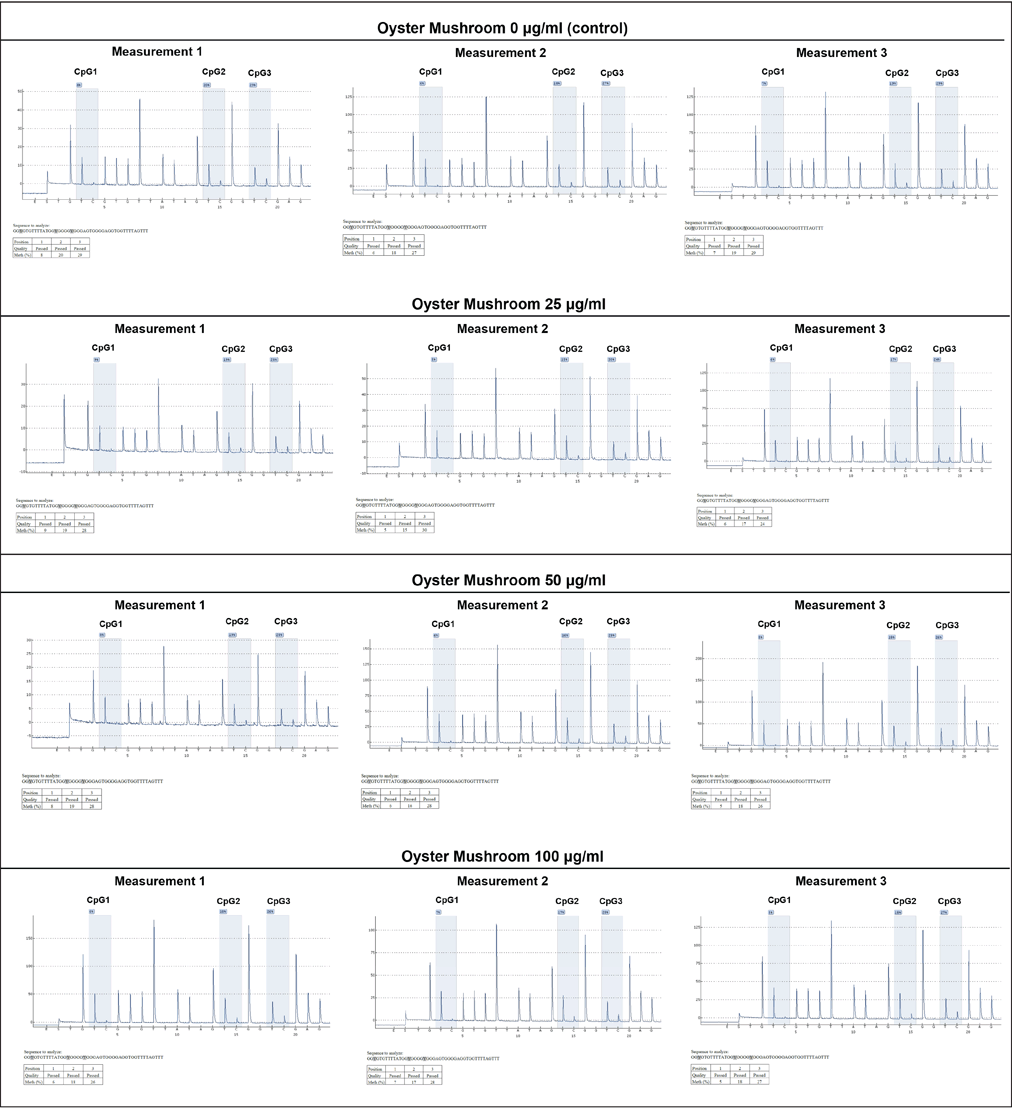 | Figure 3. DNA methylation level on Slc2a4 promoter using pyrosequencing. The experiment was conducted in triplicate (measurements 1–3) on three different CpG sites (CpG1, CpG2, and CpG3) as indicated. DNA methylation level was presented in percentages as indicated on each CpG site. X-axis: nucleotide dispensation order, Y-axis: relative light intensity. Oyster mushroom 0 µg/ml (Control): cells treated with the same concentration of ethanol without oyster mushroom extract. [Click here to view] |
A methylation level analysis was performed by analyzing the percentage of methylation at each CpG site separately and averaging the methylation percentage across all sites (Fig. 4). The pyrosequencing results showed that the percentage of methylation at CpG1 tended to decrease after treatment with the oyster mushroom ethanolic extract at concentrations of 25, 50, and 100 μg/ml, in a dose-dependent manner (Fig. 4A). After conducting normality and homogeneity tests, statistical analysis using One-Way ANOVA was performed to determine the mean differences for treatment groups analyzed for methylation at CpG1. Statistical tests revealed no significant differences between treatment groups (p > 0.05).
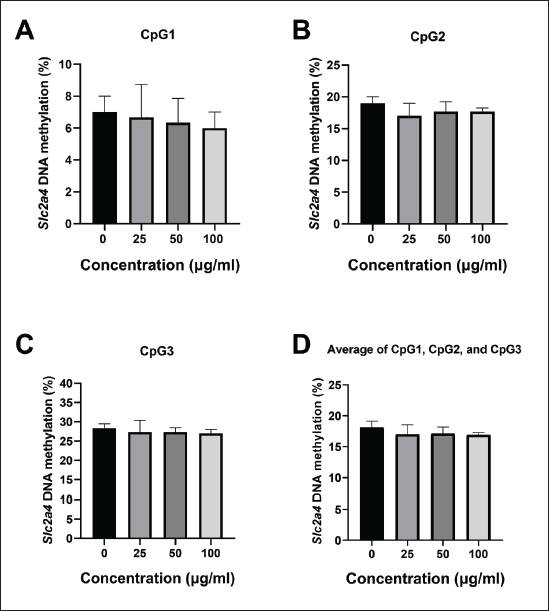 | Figure 4. Percentage of DNA methylation on Slc2a4 promoter in 3T3-L1 on day 12 following treatment with oyster mushroom ethanolic extract. The methylation level in the Slc2a4 promoter region was determined by examining the methylation percentage at (A) CpG1, (B) CpG2, (C) CpG3, and (D) average across all CpG sites. The percentage of Slc2a4 methylation was measured using pyrosequencing and calculated as the proportion of high peaks of C and T at each CpG site after bisulfite conversion. The experiment was conducted in triplicate. Kruskal-Wallis and One Way ANOVA analysis indicated no significant difference between treatment groups (p > 0.05). Data are presented as mean ± SD. 0 µg/ml (Control): cells treated with the same concentration of ethanol without the oyster mushroom extract. [Click here to view] |
The oyster mushroom ethanolic extract concentrations of 25, 50, and 100 μg/ml also tended to decrease the percentage of Slc2a4 methylation at CpG2 and CpG3 as compared to the control (0 μg/ml), although not in a dose-dependent manner (Fig. 4B and C). Statistical analysis was conducted using the non-parametric Kruskal–Wallis test after performing normality and homogeneity tests. There were no significant differences between treatment groups (p > 0.05).
DISCUSSION
This study evaluated the potential effects of an oyster mushroom ethanolic extract on lipid accumulation and the expression of Cebpa and Slc2a4 genes during 3T3-L1 adipocyte differentiation, as well as its impact on insulin sensitivity. Moreover, we explored the influence of the epigenetic process of DNA methylation on the promoter region, which was believed to play a role in gene expression regulation.
The importance of subcutaneous adipose tissue expansion in safely accommodating excess lipids has been clearly demonstrated in several studies. Kim et al. [32] stated that despite profound subcutaneous obesity due to adiponectin overexpression, mice with hyperplastic “healthy” adipose tissue still maintain insulin sensitivity comparable to that of their lean littermates [32]. The hyperplasia of adipocytes can prevent insulin resistance because it prevents adipocyte hypertrophy [7]. Hyperplasia can be a sign of the expansion of “healthy” adipocytes, marked by the accumulation of lipid droplets in the cells. We found that the oyster mushroom ethanolic extract increased lipid levels macroscopically at concentrations of 25 and 50 μg/ml. The microscopic analysis at 40× and 100× magnifications corroborated these findings, with the 50-μg/ml concentration showing the highest increase in the 3T3-L1 cell lipid levels among the groups considered (Fig. 1). Another fungus, Pleurotus citrinopileatus, has been shown to reduce serum triglycerides, cholesterol, and low-density lipoprotein [33]. This could be attributed to the reduction of FAM3A in mice, which induces adipogenesis. In the 3T3-L1 cell line, the FAM3A expression increases during adipogenesis, potentially explaining the enhanced lipid accumulation observed with the oyster mushroom extract [34].
C/EBPα is an adipogenic transcription factor encoded by the Cebpa gene, which activates the transcription of many markers expressed by mature adipocytes, such as adiponectin, IRs, and fatty acid binding protein (FABP4). The Cebpa gene activates IRs that bind to insulin on the surface of adipocytes to initiate the insulin signaling cascades [9,10,35]. Upon insulin binding, IR autophosphorylates and activates IRS proteins [35]. Phosphorylated IRS proteins recruit and activate PI3K, which leads to the generation of phosphatidylinositol-3,4,5-trisphosphate. It activates Akt, a key mediator of insulin signaling. Akt activation stimulates the translocation of GLUT4 vesicles to the cell membrane of adipocytes [35].
GLUT4 is a highly expressed protein in both adipose tissue and skeletal muscle, encoded by Slc2a4. The translocation of GLUT4 to the cell membrane facilitates glucose absorption into adipocytes [36,37]. The uptake of glucose into adipocytes induces ChREBP expression and de novo lipogenesis (the production of fatty acids from glucose) [38]. GLUT4 is also essential for maintaining overall glucose homeostasis and insulin sensitivity. Changes in the GLUT4 expression are associated with insulin resistance, a major factor in the pathogenesis of T2DM. It has been found that GLUT4 expression levels are decreased in adipose cells in insulin-resistant and pre-diabetic obese subjects, as well as in subjects with T2DM [39,40]. A study carried out by Yang et al. [41] revealed that specific adipose Slc2a4 knockout mice show secondary insulin resistance in muscle and liver through increased serum retinol-binding protein-4 levels [41]. Conversely, excessive Slc2a4 expression enhances glycemic control in diabetic mice [42]. This emphasizes the importance of the Slc2a4/GLUT4 expression in adipose tissue in body glucose control. Thus, modulating the Cebpa and Slc2a4 expression might be a potential therapeutic target for insulin resistance management and prevention in T2DM.
According to the current study, the oyster mushroom ethanolic extract increased the Cebpa and Slc2a4 gene expression as compared to that in the control cells, which were only treated with MDI stimulation, although the increase is not statistically significant (Fig. 2). It is also supported by another study that showed that Cebpa increases Slc2a4 gene expression and GLUT4 protein via ESR1 activation, following estradiol (E2) treatment during 3T3-L1 differentiation [15]. Hispolon, a phenolic acid that may be present in oyster mushrooms, is known to elevate the serum levels of 17β-estradiol or E2 [43]. Thus, it is hypothesized that increased Cebpa gene expression may occur by the hispolon bioactive.
The results of the current study also suggested that the oyster mushroom ethanolic extract increased the mRNA expression of Slc2a4 depending on the dose administered (Fig. 2B). Therefore, the oyster mushroom ethanolic extract had a direct effect on the adipose Slc2a4, leading us to hypothesize that the oyster mushroom ethanolic extract might regulate glucose homeostasis and insulin sensitivity. Aligned with the findings of previous studies, bioactive compounds contained in P. ostreatus could increase the GLUT4 expression by modulating the insulin signaling pathways. Xiong et al. [29] found that the ergosterol content in P. ostreatus acts as a hypoglycemic agent by stimulating the GLUT4 translocation and expression modulated by the PI3K/Akt and PKC pathways in L6 cells [29]. In line with a study conducted on streptozotocin-induced diabetic rats, Pleurotus ostreatus polysaccharide administration has antidiabetic effects by increasing glycogen storage via activating the GSK3 phosphorylation and the GLUT4 translocation in the muscles, reducing hyperglycemia and hyperlipidemia levels, and improving insulin resistance [28]. Other in vivo studies have also revealed that a P. ostreatus ethanolic extract significantly reduces the blood glucose levels in alloxan-induced diabetic rats [26].
Epigenetic mechanisms, particularly DNA methylation, play a critical role in regulating gene expression. DNA methylation does not modify the DNA sequence, but it can alter the expression of methylated genes. Previous research has shown that when DNA hypermethylation occurs in the gene promoter region, it is related to transcriptional repression, whereas DNA hypomethylation is associated with transcriptional activation [44,45]. The data show that an oyster mushroom ethanolic extract reduces the level of Slc2a4 methylation, although the reduction is not statistically significant. Decreases in the Slc2a4 methylation percentage were observed at the three CpG sites on the analyzed Slc2a4 promoter: CpG1, CpG2, and CpG3 (Fig. 4). The discovery of a decreased Slc2a4 methylation level supported the previous qRT-PCR results, where there was increased Slc2a4 mRNA expression with increasing doses. This finding aligned with the theory that DNA hypomethylation correlates with transcriptional activation [44]. By inducing the hypomethylation of the Slc2a4 promoter, the oyster mushroom ethanolic extract seemed to be involved in the epigenetic regulation of the Slc2a4 expression in differentiated adipocyte cell lines.
The result of a previous study suggested that DNA hypomethylation can develop as a result of decreased levels of S-adenosyl-l-methionine (SAM), increased levels of S-adenosylhomocysteine (SAH), lower SAM/SAH ratio, and decrease in the DNA methyltransferase (DNMT) activity [46–48]. The hypomethylation of the Slc2a4 promoter may also occur because of the capacity of the oyster mushroom ethanolic extract to inhibit the activity of DNMTs because of its quercetin content [48,49]. DNMTs function to catalyze DNA methylation, so the absence or inactivity of DNMTs leads to DNA demethylation and subsequently increases gene expression [50].
In this study, we only examined the Cebpa expression at the mRNA level. Another study showed that the C/EBPα expression levels were upregulated because of compensating for the lack of C/EBPβ. This compensation occurred at the Cebpa protein level, suggesting a post-transcriptional or post-translational regulatory mechanism [51]. Thus, assessing the C/EBPβ expression and C/EBPα at the protein levels is recommended. Insulin sensitivity improvement may also occur through various molecular pathways. Therefore, it is recommended to explore other genes responsible for insulin sensitivity regulation, such as genes involved in insulin signaling pathways in the adipose tissue.
We acknowledge our limitation due to the absence of mechanistic experiments to confirm that oyster mushroom extract affects Cebpa and Slc2a4 gene expression via epigenetic modulation. Therefore, we recommend exploring other epigenetic mechanisms related to gene expression regulation, including post-translational modifications of histones such as histone methylation and acetylation, non-coding RNAs, and the epigenetic regulation of Cebpa and Slc2a4. We also suggest assessing the DNMT activity to elucidate the molecular mechanisms underlying the reduction in gene methylation and investigating specific bioactive content in the oyster mushroom ethanolic extract.
Although the findings of this study did not reach statistical significance, the trends in lipid accumulation and gene expression suggest a potential modulatory effect of the oyster mushroom ethanolic extract on pathways relevant to insulin resistance. This lack of significance may be attributed to the exploratory nature of the study, with a limited sample size (e.g., gene expression analysis conducted in duplicates) that may not have fully captured the therapeutic potential of the extract. While technical precision was maintained by running each sample in parallel under consistent conditions, triplicate measurements would provide greater statistical reliability. A dose-dependent decrease in Slc2a4 promoter methylation was observed at CpG1, indicating a possible epigenetic response to the oyster mushroom extract. However, this pattern was not evident at CpG2 and CpG3. We believe this inconsistency reflects biological variability, which could influence the methylation dynamics at different CpG sites. Future studies with larger sample sizes may help clarify these relationships by better accounting for this biological variability.
Additionally, while Gapdh was used as the housekeeping gene, validating multiple reference genes would enhance normalization accuracy. Future studies should evaluate the stability of multiple reference genes under different treatment conditions to improve the robustness of gene expression analysis. Despite these recommendations, this study showed the potential use of an oyster mushroom ethanolic extract as a preventive measure against insulin resistance and to reduce the risk of T2DM.
CONCLUSION
The oyster mushroom ethanolic extract promoted lipid accumulation, upregulated the expression of Cebpa and Slc2a4, and reduced the methylation level of the Slc2a4 gene promoter. However, these effects were not statistically significant.
LIST OF ABBREVIATIONS
DMEM, Dulbecco’s modified Eagle’s medium; DMSO, Dimethyl sulfoxide; DNMTs, DNA methyltransferases; FAB4, fatty acid binding protein; FBS, fetal bovine serum; GLUT4, glucose transporter type 4; IR, insulin receptors; qRT-PCR, quantitative reverse transcription polymerase chain reaction; RB4, retinol-binding protein-4; SAH, S-adenosylhomocysteine; SAM, S-adenosyl-l-methionine; T2DM, type 2 diabetes mellitus.
ACKNOWLEDGMENT
This study was funded by Universitas Padjadjaran research grant number 1577/UN6.3.1/PT.00/2024 for EFA and Ministry of Education, Culture, Research and Technology of Indonesia research grant number 3866/UN6.3.1/PT.00/2024 for EFA.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Ethical approvals details are given in the ‘Materials and Methods’ section.
DATA AVAILABILITY
All the data are available with the corresponding author (EFA) and shall be provided upon request.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declare that they have not used artificial intelligence (Al)-tools for writing and editing the manuscript, and no images were manipulated using Al.
REFERENCES
1. Standl E, Khunti K, Hansen TB, Schnell O. The global epidemics of diabetes in the 21st century: current situation and perspectives. Eur J Prev Cardiol. 2019 Dec;26(2_suppl):7–14. CrossRef
2. WHO. Diabetes [Internet]. Geneva, Switzerland: WHO; 2024 [cited 2024 Apr 2]. Available from: https://www.who.int/news-room/fact-sheets/detail/diabetes
3. Wahidin M, Achadi A, Besral B, Kosen S, Nadjib M, Nurwahyuni A, et al. Projection of diabetes morbidity and mortality till 2045 in Indonesia based on risk factors and NCD prevention and control programs. Sci Rep. 2024 Mar 5;14:5424. CrossRef
4. American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010 Jan;33(Suppl 1):S62–9. CrossRef
5. Freeman AM, Acevedo LA, Pennings N. Insulin resistance. Treasure Island, FL: StatPearls Publishing; 2024 [cited 2024 Apr 2]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK507839/
6. Carvalho E, Kotani K, Peroni OD, Kahn BB. Adipose-specific overexpression of GLUT4 reverses insulin resistance and diabetes in mice lacking GLUT4 selectively in muscle. Am J Physiol-Endocrinol Metab. 2005 Oct;289(4):E551–61. CrossRef
7. Gustafson B, Hedjazifar S, Gogg S, Hammarstedt A, Smith U. Insulin resistance and impaired adipogenesis. Trends Endocrinol Metab. 2015 Apr 1;26(4):193–200. CrossRef
8. Reitman ML, Arioglu E, Gavrilova O, Taylor SI. Lipoatrophy revisited. Trends Endocrinol Metab TEM. 2000 Dec;11(10):410–6. CrossRef
9. Ali AT, Hochfeld WE, Myburgh R, Pepper MS. Adipocyte and adipogenesis. Eur J Cell Biol. 2013 Jun 1;92(6):229–36. CrossRef
10. Zhao J, Zhou A, Qi W. The potential to fight obesity with adipogenesis modulating compounds. Int J Mol Sci. 2022 Feb 19;23(4):2299. CrossRef
11. Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 2019 Apr;20(4):242–58. CrossRef
12. Li X, Zeng S, Chen L, Zhang Y, Li X, Zhang B, et al. An intronic enhancer of Cebpa regulates adipocyte differentiation and adipose tissue development via long-range loop formation. Cell Prolif. 2023 Oct 31;57(3):e13552. CrossRef
13. Matulewicz N, Stefanowicz M, Niko?ajuk A, Karczewska-Kupczewska M. Markers of adipogenesis, but not inflammation, in adipose tissue are independently related to insulin sensitivity. J Clin Endocrinol Metab. 2017 Aug 1;102(8):3040–9. CrossRef
14. Boughanem H, Cabrera-Mulero A, Millán-Gómez M, Garrido-Sánchez L, Cardona F, Tinahones FJ, et al. Transcriptional analysis of FOXO1, C/EBP-α and PPAR-γ2 genes and their association with obesity-related insulin resistance. Genes. 2019 Sep 12;10(9):706. CrossRef
15. Fatima LA, Campello RS, Barreto-Andrade JN, Passarelli M, Santos RS, Clegg DJ, et al. Estradiol stimulates adipogenesis and Slc2a4/GLUT4 expression via ESR1-mediated activation of CEBPA. Mol Cell Endocrinol. 2019 Dec 1;498:110447. CrossRef
16. Carvalho E, Jansson PA, Nagaev I, Wenthzel AM, Smith U. Insulin resistance with low cellular IRS-1 expression is also associated with low GLUT4 expression and impaired insulin-stimulated glucose transport. FASEB J Off Publ Fed Am Soc Exp Biol. 2001 Apr;15(6):1101–3. CrossRef
17. Carvalho E, Jansson PA, Axelsen M, Eriksson J, Huang X, Groop L, et al. Low cellular IRS 1 gene and protein expression predict insulin resistance and NIDDM. FASEB J. 2000 Jan 1;13:2173–8. CrossRef
18. Weinhold B. Epigenetics: the science of change. Environ Health Perspect. 2006 Mar;114(3):A160–7. CrossRef
19. Li Y, Tollefsbol TO. DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol Biol Clifton NJ. 2011;791:11–21. CrossRef
20. Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–13. CrossRef
21. Hasan MM, Ahmed QU, Soad SZM, Latip J, Taher M, Syafiq TMF, et al. Flavonoids from Tetracera indica Merr. induce adipogenesis and exert glucose uptake activities in 3T3-L1 adipocyte cells. BMC Complement Altern Med. 2017 Aug 30;17(1):431. CrossRef
22. Bhushan A, Kulshreshtha M. The medicinal mushroom Agaricus bisporus: review of phytopharmacology and potential role in the treatment of various diseases. J Nat Sci Med. 2018 Mar 1;1:18. CrossRef
23. Kumar K. Nutraceutical potential and processing aspects of oyster mushrooms (Pleurotus species). Curr Nutr Food Sci. 2020;16(1):3–14. CrossRef
24. de Arruda EHP, de Reis MAB, Marin L, Muller LA, Damazo AS, Gerenutti M, et al. Investigation into the intake of edible mushroom Pleurotus ostreatus (aqueous extract oyster mushroom) on biochemical indices of female wistar Rats. Am J Plant Sci. 2023 Feb 9;14(2):177–90. CrossRef
25. Ravi B, Renitta RE, Prabha ML, Issac R, Naidu S. Evaluation of antidiabetic potential of oyster mushroom (Pleurotus ostreatus) in alloxan-induced diabetic mice. Immunopharmacol Immunotoxicol. 2013 Feb;35(1):101–9. CrossRef
26. Choudhury B, Rahman T, Kakon A, Hoque N, Akhtaruzzaman M, Begum M, et al. Effects of Pleurotus ostreatus on blood pressure and glycemic status of hypertensive diabetic male volunteers. J Med Biochem. 2013 Jan 13;6:5–10. CrossRef
27. Asrafuzzaman M, Rahman MM, Mandal M, Marjuque M, Bhowmik A, Rokeya B, et al. Oyster mushroom functions as an anti-hyperglycaemic through phosphorylation of AMPK and increased expression of GLUT4 in type 2 diabetic model rats. J Taibah Univ Med Sci. 2018 Oct;13(5):465–71. CrossRef
28. Zhang Y, Hu T, Zhou H, Zhang Y, Jin G, Yang Y. Antidiabetic effect of polysaccharides from Pleurotus ostreatus in streptozotocin-induced diabetic rats. Int J Biol Macromol. 2016 Feb;83:126–32. CrossRef
29. Xiong M, Huang Y, Liu Y, Huang M, Song G, Ming Q, et al. Antidiabetic activity of ergosterol from Pleurotus ostreatus in KK-Ay mice with spontaneous type 2 diabetes mellitus. Mol Nutr Food Res. 2018 Feb;62(3):444. CrossRef
30. Shao HY, Hsu HY, Wu KS, Hee SW, Chuang LM, Yeh JI. Prolonged induction activates Cebpα independent adipogenesis in NIH/3T3 cells. PLoS One. 2013 Jan 10;8(1):e51459. CrossRef
31. Ariyanto EF, Shalannandia WA, Lantika UA, Fakih TM, Ramadhan DSF, Gumilar AN, et al. Anthocyanin-containing purple sweet potato (Ipomoea batatas L.) synbiotic yogurt inhibited 3T3-L1 adipogenesis by suppressing white adipocyte-specific genes. J Exp Pharmacol. 2023 May 22;15:217–30. CrossRef
32. Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest. 2007 Sep 4;117(9):2621–37. CrossRef
33. Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006 Dec 14;444(7121):840–6. CrossRef
34. Chi Y, Li J, Li N, Chen Z, Ma L, Peng W, et al. FAM3A enhances adipogenesis of 3T3-L1 preadipocytes via activation of ATP-P2 receptor-Akt signaling pathway. Oncotarget. 2017 May 3;8(28):45862–73. CrossRef
35. Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014 Jan;6(1):a009191. CrossRef
36. Uldry M, Thorens B. The SLC2 family of facilitated hexose and polyol transporters. Pflugers Arch. 2004 Feb;447(5):480–9. CrossRef
37. Britsemmer JH, Krause C, Taege N, Geißler C, Lopez-Alcantara N, Schmidtke L, et al. Fatty Acid induced hypermethylation in the Slc2a4 gene in visceral adipose tissue is associated to insulin-resistance and obesity. Int J Mol Sci. 2023 Mar 29;24(7):6417. CrossRef
38. Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J Clin Invest. 2006 Jul 3;116(7):1767–75. CrossRef
39. Corrêa LH, Heyn GS, Magalhaes KG. The impact of the adipose organ plasticity on inflammation and cancer progression. Cells. 2019 Jul;8(7):662. CrossRef
40. Cignarelli A, Genchi VA, Perrini S, Natalicchio A, Laviola L, Giorgino F. Insulin and insulin receptors in adipose tissue development. Int J Mol Sci. 2019 Feb 11;20(3):759. CrossRef
41. Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005 Jul;436(7049):356–62. CrossRef
42. Michael Gibbs E, Stock JL, McCoid SC, Stukenbrok H, Pessin JE, Stevenson RW, et al. Glycemic improvement in diabetic db/db mice by overexpression of the human insulin-regulatable glucose transporter (GLUT4). J Clin Investig. 1995 Apr 1;95(4):1512–8. CrossRef
43. Wang J, Chen B, Hu F, Zou X, Yu H, Wang J, et al. Effect of hispolon from Phellinus lonicerinus (Agaricomycetes) on estrogen receptors, aromatase, and cyclooxygenase II in MCF-7 breast cancer cells. Int J Med Mushrooms. 2017;19(3):233–42. CrossRef
44. Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011 May 15;25(10):1010–22. CrossRef
45. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012 May 29;13(7):484–92. CrossRef
46. Ghoshal K, Li X, Datta J, Bai S, Pogribny I, Pogribny M, et al. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. J Nutr. 2006 Jun;136(6):1522–7. CrossRef
47. Wainfan E, Poirier LA. Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res. 1992 Apr 1;52(7 Suppl):2071s–7s.
48. Devi PV, Islam J, Narzary P, Sharma D, Sultana F. Bioactive compounds, nutraceutical values and its application in food product development of oyster mushroom. J Future Foods. 2024 Dec 1;4(4):335–42. CrossRef
49. Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005 Oct;68(4):1018–30. CrossRef
50. Hervouet E, Peixoto P, Delage-Mourroux R, Boyer-Guittaut M, Cartron PF. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin Epigenetics. 2018 Feb 9;10(1):17. CrossRef
51. Lau KH, Waldhart AN, Dykstra H, Avequin T, Wu N. PPARγ and C/EBPα response to acute cold stress in brown adipose tissue. iScience. 2023 Jan 1;26(1):105848–8. CrossRef