INTRODUCTION
Fungi represent one of the most prevalent kingdoms of organisms on earth with more than 12,000 reported species. The updated estimate of the number of undiscovered species falls within the range of 2.2 to 3.8 million [1,2]. These microorganisms have vast and varied economic and ecological impacts, ranging from the manufacture of life-saving pharmaceuticals and the fermentation industry [3,4] to human and animal infections, as well as mycotoxin-induced poisoning of crops, livestock, and food [5].
The Aspergillus genus is considered the most widespread fungal genus worldwide with more than 330 accepted species [6]. These species are widely found in different natural habitats and play crucial roles in the decomposition of organic materials, while some species produce a variety of mycotoxins that lead to destructive rots in agricultural and food products [7]. These species can produce a variety of secondary metabolites, including terpenoids, alkaloids, glycosides, peptides, polyketides, and steroids. These metabolites possess promising activities such as ovicidal, antifungal, cytotoxic, antibacterial, nematicidal, radical scavenging, and insect growth-regulating properties [14–22]. Some of them have potential applications in the medicinal and agricultural sectors. For example, lovastatin and simvastatin, well-known cholesterol-lowering medications, were biosynthesized and synthetically derived from Aspergillus terreus, respectively. They have been shown to promote HMG-CoA reductase inhibition and are utilized in clinical settings for treating cardiovascular disease and hypercholesterolemia [23].
Some species such as Aspergillus sclerotiorum and Aspergillus ochraceus are of economic importance because of their role in the biochemical transformation of phenazines, steroids, and alkaloids [15,19,24]. Besides, Aspergillus melleus and Aspergillus ochraceus are potential producers of proteolytic enzymes and various bio-metabolites [15,19,24]. On the other hand, A. ochraceus and A. sclerotiorum have been reported as human and animal pathogens that cause antromycosis, onychomycosis, allergic bronchopulmonary aspergillosis, and otomycosis in humans, as well as mycotic placentitis in the cow [24].
Unlike other Aspergillus species, Aspergillus ochraceopetaliformis Bat. & Maia, a rarely studied fungus, has not yet been recognized as a well-known human pathogen. It had been identified to cause infection of a healthy woman’s toenails that had been treated effectively by ciclopiroxolamine plus terbinafine [24,25]. Aspergillus ochraceopetaliformis was identified as a soft-rot fungus that degrades archeological wood at the Al-Aqsa Mosque [7]. Recently, it was isolated from Setipinna phasa (Shukti) dried fish [26]. Ngo et al. noted that A. ochraceopetaliformis produced high levels of exopolysaccharide (9.89 g/l) that are major contributors to significant physical, chemical, and aesthetical alterations and leaching of elements from glass surfaces [27]. On the other hand, this fungus produces diverse classes of secondary metabolites: pyran derivatives, cyclopentenones, lactones, polyketides, anthraquinones, biphenyl ethers, sesquiterpenoids, alkaloids, isocoumarins, and cyclopeptides with significant pharmacological properties.
Despite many studies investigating various Aspergillus species, there is a lack of review that combines the pharmacological properties, mechanism of actions, and biosynthesis pathways of A. ochraceopetaliformis metabolites.
Therefore, the current review aims to provide a comprehensive discussion of A. ochraceopetaliformis, including its secondary metabolites and their biological properties. Additionally, the biosynthetic pathways of these metabolites were highlighted. These compounds were classified based on their chemical skeletons. This work is the first to thoroughly discuss the biosynthetic pathways and bio-activities of A. ochraceopetaliformis metabolites, highlighting this species’ potential as a source of new bioactive compounds, particularly for the treatment of viral infections, inflammation, and cancer.
SEARCH METHODOLOGY
A literature search using databases, including PubMed, Scopus, Web of Science, and Google Scholar was done to compile all reported studies on A. ochraceopetaliformis, its secondary metabolites, and biological activities. The search employed keywords such as “Aspergillus ochraceopetaliformis + secondary metabolites” OR “Aspergillus ochraceopetaliformis + biosynthetic pathways”, “Aspergillus ochraceopetaliformis + biological activities”. The studies reported on isolation, identification, biosynthesis, and biological properties of A. ochraceopetaliformis metabolites were included, while publications unrelated to these areas on A. ochraceopetaliformis were excluded.
ASPERGILLUS OCHRACEOPETALIFORMIS METABOLITES AND THEIR BIOACTIVITIES
Pyran derivatives
α-Pyrone mero-sesquiterpenoids have an angular tetracyclic skeleton, consisting of α-pyrone and highly oxygenated trans-decalin substructures. These compounds have been previously reported from Penicillium and Aspergillus species [28].
Wang et al. separated new α-pyrone mero-sesquiterpenoids: ochraceopones A-E (1–5) and isoasteltoxin (6), along with 7 and 8 from the ethyl acetate (EtOAc) extract of nutrient-deprived medium of Antarctic soil-derived A. ochraceopetaliformis SCSIO-05702 collected near the Great Wall station using SiO2/Sephadex LH-20 CC/high-performance liquid chromatography (HPLC) [8] (Table 1).
 | Table 1. List of pyran derivatives isolated from Aspergillus ochraceopetaliformis. [Click here to view] |
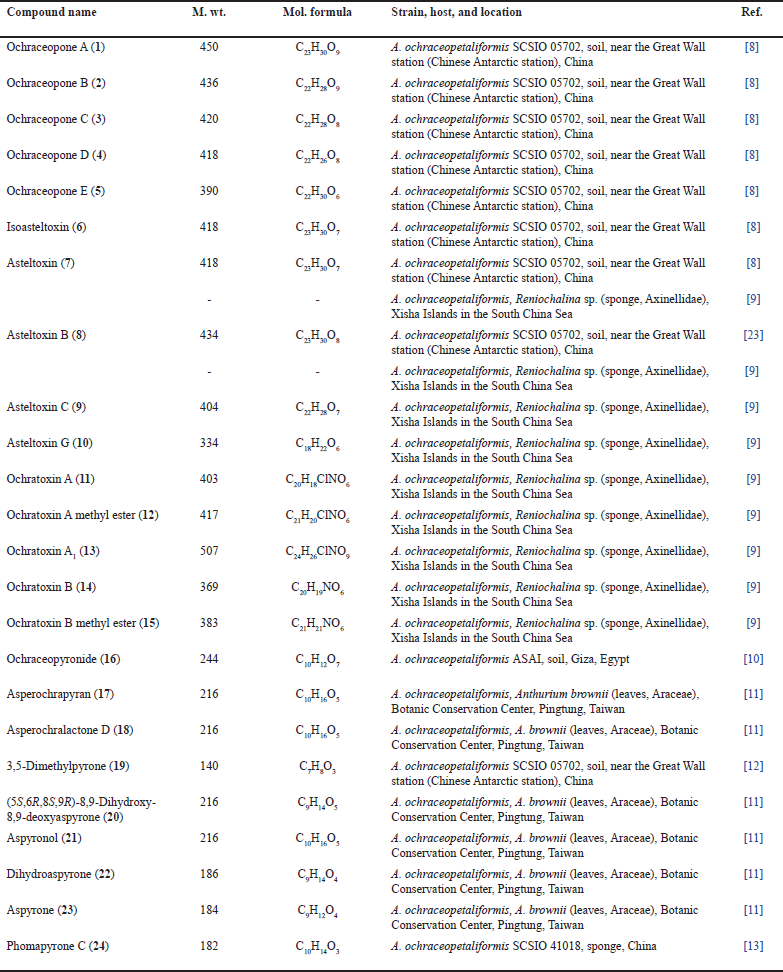
Ochraceopones A–D (1–4) are α-pyrone mero-sesquiterpenoids, having a tetracyclic linear carbon skeleton that differs at position 6, possessing CH-OCH3, CH-OH, and CH2, respectively (Fig. 1). The C-7 methine and C-6 oxymethine in 2 are replaced by C-6-C7 double bond in 4 [8]. Their 6R/7S/8S/9R/10R/11R, 12R/6R/7S/8S/9R/10R/11R/12R, 7R/8S/9R/10R/11R/12R, and 8S/9R/10R/11R/12R configurations, respectively, were assigned based on nuclear overhauser effect spectroscopy (NOESY)/CD/Xray analyses.
 | Figure 1. Chemical structures of pyrones derivatives (1–8) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
Compound 5 is 7R/8S/9R/11S/12R configured and has different connectivity of decalin unit from 3. Compound 6 possesses α-pyrone and 2,8-dioxabicyclo[3.3.0]octane units as 7 with Z-geometry of C-12-C-11 double bond. Compounds 1−5 are proposed to be generated via mevalonate/polyketide pathways. Trans-farnesyl pyrophosphate (FPP) is formed through a mevalonate pathway that combines with malonyl-CoA and acetyl-CoA to produce an intermediate A (Fig. 2). The tetracyclic core is formed from this intermediate through epoxidation, polyene cyclization, and retro-aldol/aldol rearrangement. Compounds 1–5 are created through oxidation, epoxidation, cyclization, retro-aldol/aldol rearrangement, dehydration, and methylation [8]. Compounds 1−8 were tested for anti-H1N1 and H3N2 activities. Compounds 6 and 7 displayed powerful antiviral capacities against the H3N2 and H1N1 influenza viruses (IC50s 0.66/0.23 and 0.84/0.54 µM, respectively) with anti-H1N1 selectivity indexes (SIs) 2.35 and 0.44, respectively, compared to Tamiflu (IC50s 18.5 and 16.9 nM, respectively) in the cytopathic effect inhibition assay. These findings suggested that the polyene chain Δ11 double bond geometry contributes to the anti-H1N1 efficacy [8].
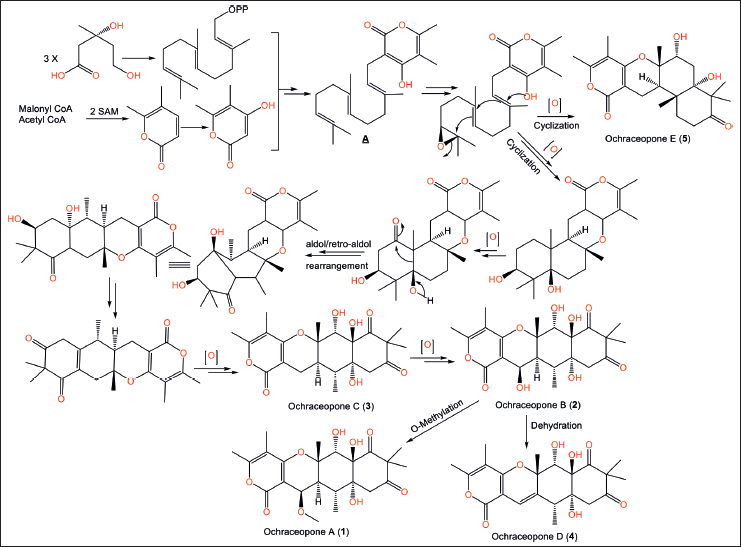 | Figure 2. Biosynthetic pathways of ochraceopones A–E (1–5) [8] . [Click here to view] |
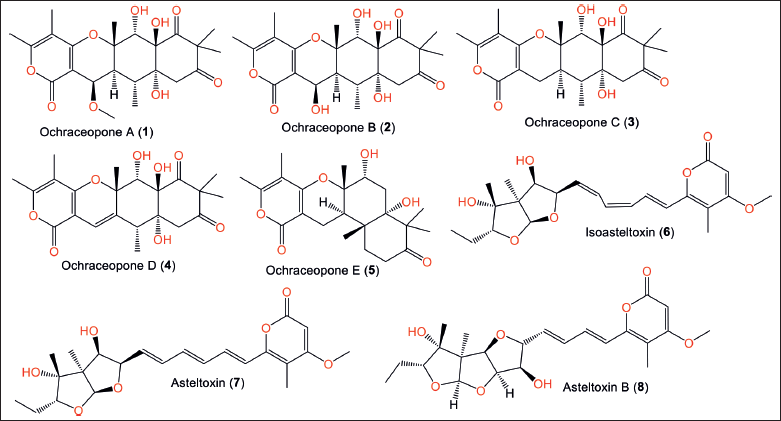
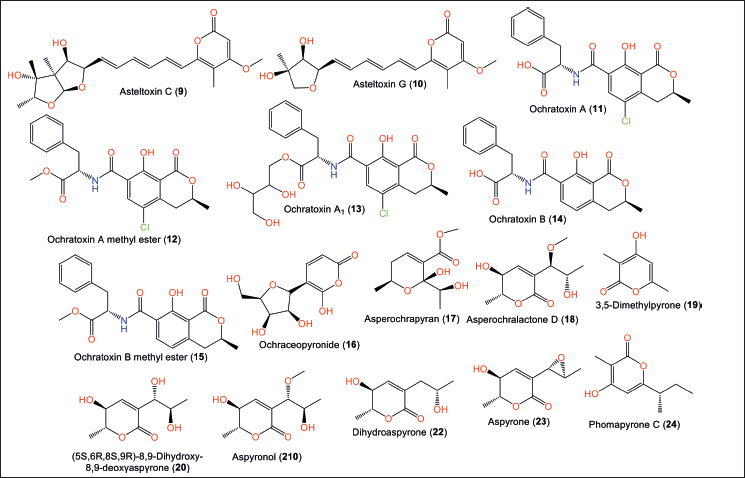
Compounds 10 and 13 are new trienic α-pyrone and ochratoxin derivatives, respectively, that were reported from Reniochalina sp. sponge-associated A. ochraceopetaliformis collected from the Xisha Islands/South China Sea, in addition to 7–9, 11, 12, 14, and 15 using silica gel column chromatography/ODS/HPLC [9]. Compound 10 has a pyrone-triene core attached to a substituted tetrahydrofuran moiety. While 13 (αD-57.5) was like 11 ([α]D-66) with an additional butane-1,2,3,4-tetraol (Fig. 3).
 | Figure 3. Chemical structures of pyrone derivatives (9–24) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
Compounds 13–15 displayed a notable reduction in the tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) induced lipopolysaccharide (LPS) expression in human monocytic cell line (THP-1) cells (%inhibition 67.7%, 72.8%, and 72.9% for TNF-α and 74.4%, 91.6%, and 89.7%, respectively, for IL-6) without cytotoxicity after 24 hours treatment [9]
Mitani et al. identified compound 7 as a potential inhibitor of mitochondrial Adenosine triphosphate (ATP) synthase and extracellular vesicles (EVs) [31]. It suppressed EV secretion without causing mitochondrial damage by reducing ATP levels and triggering 5’ adenosine monophosphate-activated protein kinase-mediated mechanistic target of rapamycin C1 inactivation, leading to nuclear translocation of microphthalmia/transcription factor E transcription factors and enhancing lysosomal gene expression and activation. It revealed increased lysosomes and reduced multivesicular bodies (MVBs) and EV levels, highlighting 7 as a novel EV inhibitor that influences MVB dynamics [31].
Asmaey et al. identified 16 as a rare and new α-pyrone-C-glycoside derivative from A. ochraceopetaliformis collected from Giza province/Egypt. This compound was assigned as 6-OH-2-pyrone-5-C-lyxofuranoside, which has 6-OH-a-pyrone with C-5-linked pentofuranoside moiety. Compound 16 exhibited weak antimicrobial potential versus different microbes [10].
Hu et al. [11] reported that Anthurium brownii-harboring A. ochraceopetaliformis EtOAc extract possessed marked anti-inflammatory capacity by suppressing elastase release and superoxide anion production (%inhibition 107% and 103 %, respectively; Conc. 10 μg/ml) [11]. Its bio-guided separation led to the identification of new pyran derivatives: 17 and 18, along with 20–23. Compounds 17 and 18’s 2R/6S/9S and 5S/6R/8R/9S configurations, respectively, were assigned based on electronic circular dichroism (ECD) analysis. Compound 23 revealed anti-inflammatory potential (% inhibition 29%, Conc. 10 μM) by inhibiting the formation of superoxide anion in neutrophils stimulated by N-formyl-L-methionyl-L-leucylL-phenylalanine [11].
Isocoumarins
Compound 25 was reported to have anti-inflammation potential through inhibition of elastase release and superoxide anion production (% inhibition 34 % and 26 %, respectively) [11]. Compounds 26–28 were reported by Wang et al. from a nutrient-rich agitated fermentation medium of SCSIO-05702 strain [12]. Compounds 26 and 27 were found to suppress LPS-caused nitric oxide (NO) production and lessened TNF-α, IL-6, and monocyte chemoattractant protein-1 (MCP-1) levels in RAW 264.7 macrophages in the cell counting kit-8 (CCK-8) assay [29]. The new isocoumarin, 29 and its related analog 30 were separated from Tethya wilhelma-harboring A. ochraceopetaliformis collected from Nanao Island/Shantou/Guangdong Province (Fig. 4). Compound 29 demonstrated cytotoxic efficacy on the B16 cell line (IC50 72.5 µM) in the MTT assay [30].
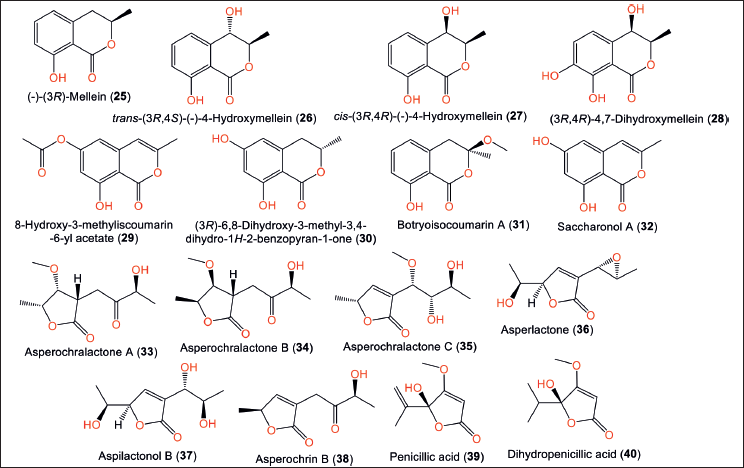 | Figure 4. Chemical structures of isocoumarin derivatives (25–32) and lactones (33–40) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
Lactones
Asperochralactones A–C (33−35), in addition to compounds 36–39 were obtained from A. ochraceopetaliformis associated with A. brownii leaves. Compounds 33−35 are new polyketides γ-lactone moiety that have 3S/4R/5R/9S, 3S/4S/5S/9S, 5R/7S/8S/9S, and 5S/6R/8R/9S, respectively (Table 2) [11].
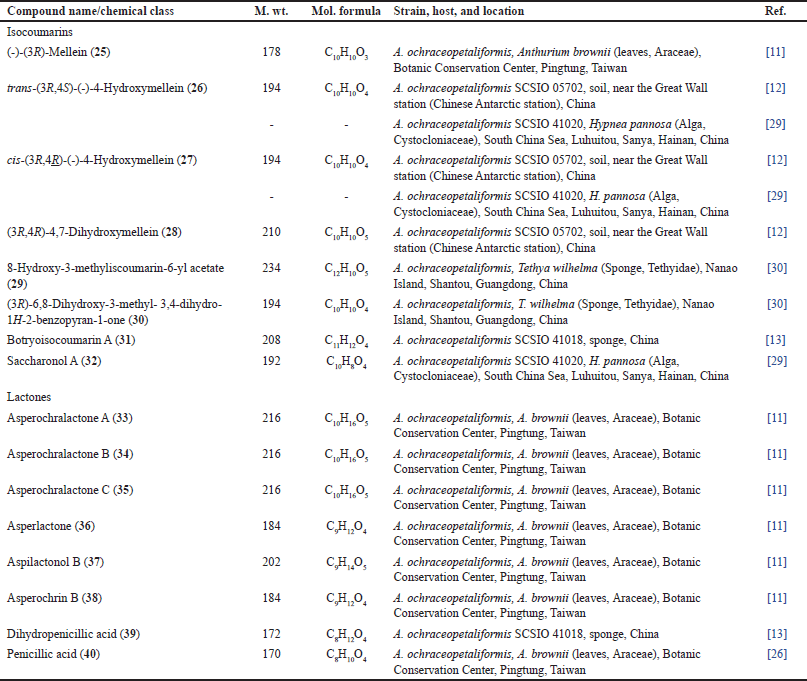 | Table 2. List of isocoumarins and lactones isolated from Aspergillus ochraceopetaliformis. [Click here to view] |
Their anti-inflammation test using fMLP-stimulated human neutrophils revealed that 36 inhibited superoxide anion formation (Conc. 10 μM; % inhibition 30%); however, compounds 36, 37, and 39 displayed elastase release inhibitory capacities (25%, 38% and 25%, respectively). Also, compounds 36 and 39 were cytotoxic against the HepG2 cancer cell line (IC50s 42.9 and 32.9 μM, respectively) [11]. Compounds 17, 18, 20, 21, 23, 33−35, and 38 have 3-oxobutanoic acid with 3-oxopentanoic or 3,5-dioxohexanoic acid that were proposed to be biosynthesized via polyketide synthase (PKS) pathway through series of reactions, involving cyclization, condenation, decarbonation, dehydration, methylation, reduction, and oxidation [11] (Fig. 5).
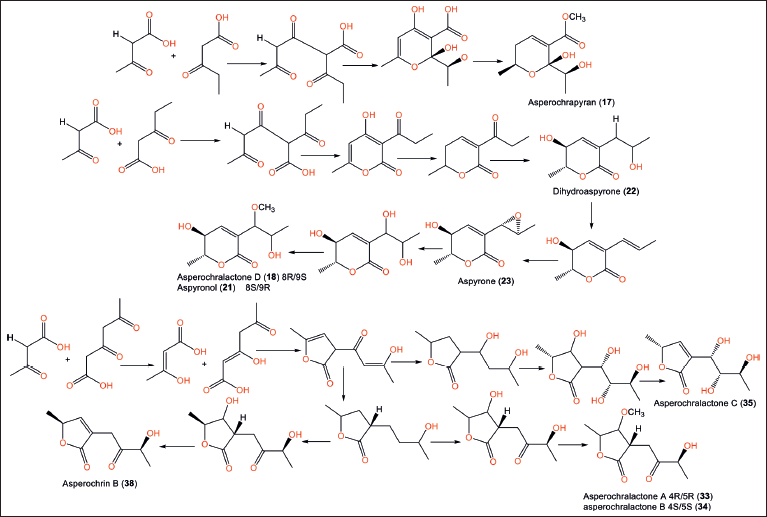 | Figure 5. Biosynthetic pathways of 17, 18, 21−23, 33−35, and 38 [11] . [Click here to view] |
Sesquiterpenoids
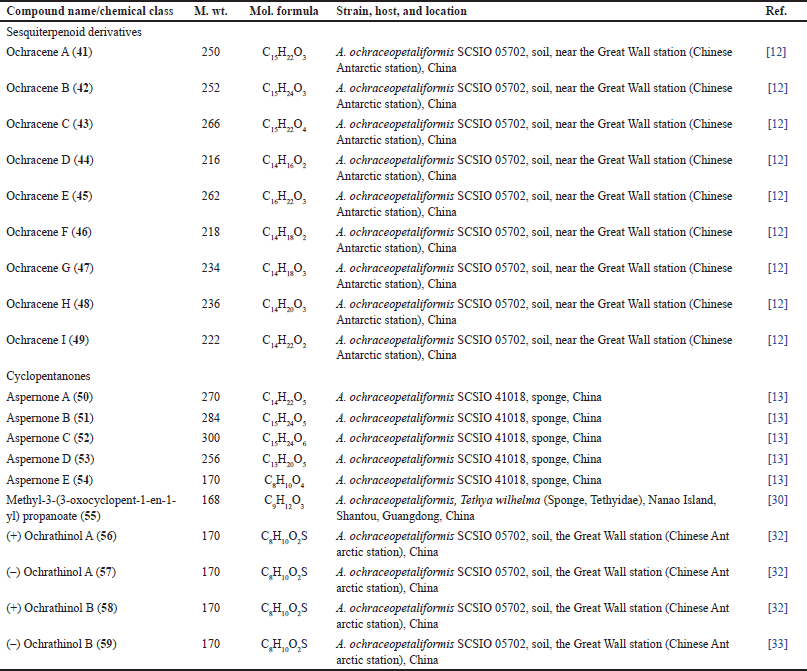
Humulane-type sesquiterpenoids, a rare class of compounds with a distinctive 11-membered ring were reported from mushrooms, plants, and liverworts, while they are uncommonly found in actinomycetes or fungi [33]. A study by Wang et al. reported that the culture condition greatly affected the type of biosynthesized metabolites. Changing the static fermentation of A. ochraceopetaliformis SCSIO 05702 in a limited-nutrient culture medium to a nutrient-rich culture with agitated fermentation resulted in the production of 41–49, new humulane sesquiterpenoids that were separated using SiO2/reversed phase-18 column chromatography (RP-18 CC)/HPLC and elucidated based on spectral/ECD analyzes and Mosher’s method. These compounds possess unprecedented carbon frameworks with ring cleavage, methyl migration, and carbon loss (Fig. 6, Table 3). The 9S/12S, 9S/12S, and 8R configurations for 44/45, 46/49, and 48/49 were assured based on specific rotation, ECD, and Mosher’s method, respectively. For instance, 42 and 43 belong to 8,9-seco-humulane sesquiterpenoids.
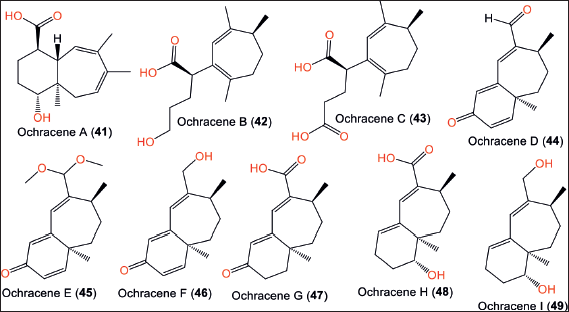 | Figure 6. Chemical structures of sesquiterpenoids (41–49) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
 | Table 3. List of sesquiterpenoids and cyclopentanones isolated from Aspergillus ochraceopetaliformis. [Click here to view] |
Compounds 42 and 43 exhibited inhibitory effectiveness on NO formation induced by LPS in RAW 264.7 mouse macrophage cell lines (IC50s 14.6 and 18.3 µM, respectively), compared to dexamethasone (IC50 2.5 µM) in cell culture and viability assay [12]. These compounds possessed no antiviral or cytotoxic activities (up to 100 µM) [12].
Biosynthetically, compounds 41–49 are generated from the humulane skeleton. This skeleton is obtained from the enzymatic cyclization of FPP (Fig. 7). A 6/7 carbon ring framework is formed by a bond between C-9 and C-4. Consequently, 41–49 are produced via a sequence of reactions, including methyl migration, oxidation, ring cleavage, and carbon loss [12].
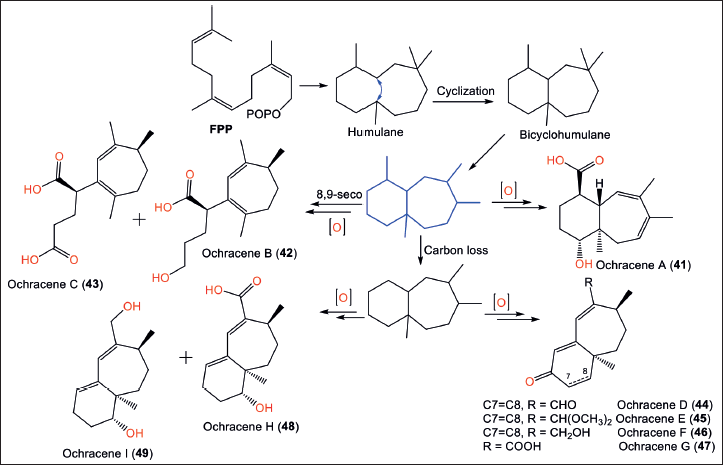 | Figure 7. Biosynthetic pathways of ochracene A−I (41–49) from humulane [12] [Click here to view] |
Cyclopentenones
From sponge-derived SCSIO 41018 strain, aspernones A–E (50–54), new cyclopentenone analogs that differ in configuration and side chain were identified using spectral/ECD/Xray analyzes (Fig. 8). Compounds 50–54 feature 3,5-dihydroxy-cyclopentenone skeleton, while 54 has hydroxylated C-2 and methyl at C-4. Their configurations were assigned as 5S/6S/21R, 5R/2`R, 5R/2`R, 4R/5R and 2S, respectively, based on ECD and Xray analyzes [13].
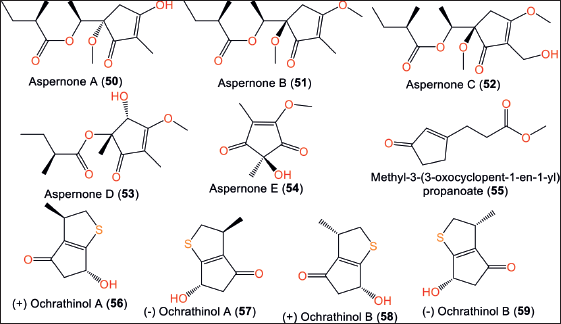 | Figure 8. Chemical structures of cyclopentanones (50–59) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
Compounds 56–59 are undescribed sulfur-containing racemates that were separated from Antarctic soil-derived A. ochraceopetaliformis SCSIO 05702 by SiO2/Sephadex LH-20 CC/HPLC/Chiral HPLC and elucidated by spectral/chiral-phase HPLC/X-ray analyses and quantum ECD calculations [32]. These enantiomers were separated by HPLC with FLM chiral-ND (2)-RH chiral column into (+) A (56), (–) A (57), (+) B (58), and (–) B (59). These compounds feature a novel 3-methylhexahydro-2H-cyclopenta [b]thiophene framework with 3R/7R, 3S/7S, 3S/7R, and 3R/7S configurations, respectively. The racemic mixture 56/57 repressed IL-6, IL-1β, and TNF-α release produced by LPS (Conc. 10 μM) and raised the unbalanced NADH/NAD+ ratio in RAW264.7 macrophages [33].
Alkaloids
In 2021, Park et al. isolated 60 from marine-derived A. ochraceopetaliformis, which is a novel sulfonated diphenyletheraminol-amino acid guanidinium salt, along with its component 61 [1-(aminoiminomethyl)-prolinol, N,N-dimethylvaline ester] that were identified by spectral/chemical methods (Fig. 9; Table 4) [34].
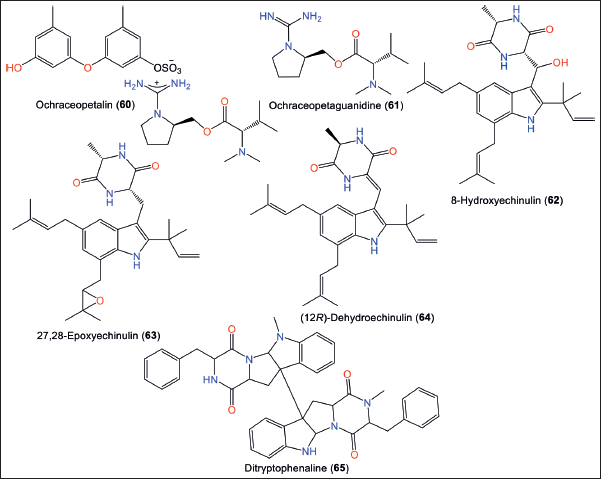 | Figure 9. Chemical structures of alkaloids (60–65) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
 | Table 4. List of alkaloids and peptides isolated from Aspergillus ochraceopetaliformis. [Click here to view] |
The salt nature of 60 was established by a set of pH-based degradation reactions. Compound 60 was proposed to originate from mixed-biogenetic salt produced from amino acid and polyketide pathways. Compound 60 had notable cytotoxic potential against A-549 and K-562 (IC50 6.8 and 9.5 µM, respectively), while 60 was weakly active in comparison to doxorubicin (IC50 0.90 and 0.72, respectively). On the other side, compounds 60 and 61 displayed no antimicrobial or sortase A (SrtA) inhibitory activities [34].
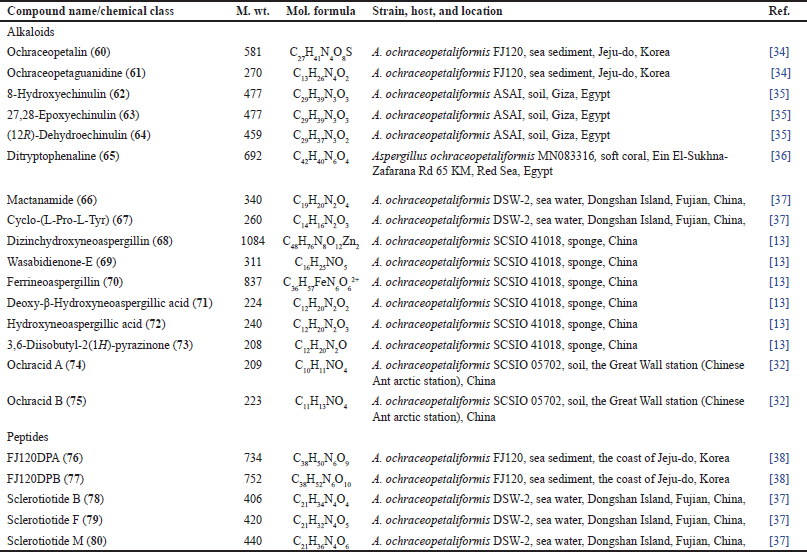
New diketopiperazine alkaloids: 62–64 were reported by Asmaey et al. from soil-derived A. ochraceopetaliformis that were assigned using spectral/Marfey’s analyses and optical rotation measurement. These compounds showed antimicrobial properties against different bacterial and fungal strains [35]. From the EtOAc extract of reef coral-associated MN083316 strain, 65 was purified that revealed marked antioxidant and antimicrobial activities, in addition to cytotoxic capacity on HEPG2 and MCF-7 (IC50s 7.6 and 5.8, respectively) [36]. Additionally, 66 and 67 were reported from seawater-derived stains that showed weak cytotoxic potential versus HPAC and BXPC-3 cell lines [37].
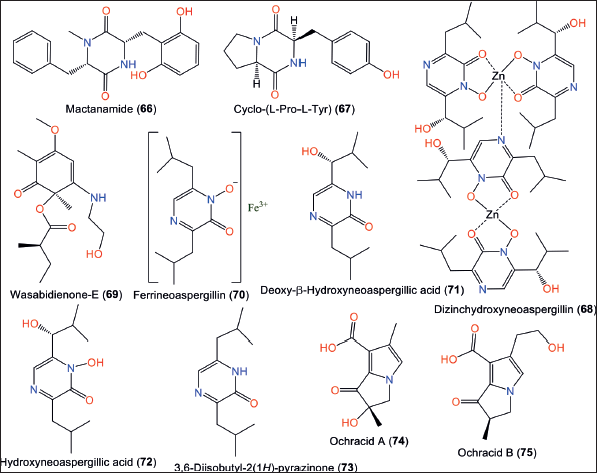
Compound 68 a new zinc complex dimer, along with 69–73 were separated by Guo et al. from sponge-associated SCSIO 41018 strain using SiO2/Sephadex LH-20/RP-18 CC/HPLC and assigned by spectral/ECD/X-ray analyses [13]. Compound 68 was identified as a zinc complex of 72, which has a Zn atom connected N-4 by a coordinate bond (Fig. 10). These compounds were assessed for their antimicrobial and cytotoxic activities. Compounds 68 and 72 demonstrated potent activities versus Enterococcus faecalis and Acinetobacter baumannii (minimum inhibitory concentrations [MICs] 0.9/0.45 and 0.9/0.45 μg/ml, respectively), compared to ampicillin and gentamicin (MICs 0.04 and 0.33 μg/ml, respectively). Further, 68 was moderately cytotoxic against the SGC-7901, human BEL-7402, and K-562 cell lines (IC50s 12.88–15.83 μM), compared to paclitaxel (0.76–1.44 μM) [13].
 | Figure 10. Chemical structures of alkaloids (66–75) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
Unprecedented pyrrolizidine alkaloids: 74 and 75 were separated from SCSIO 05702 strain derived from Antarctic soil using SiO2/Sephadex LH-20 CC/HPLC and elucidated by spectral/ECD/Xray analyses. These are bicyclic 7R-configured compounds with pyrrolidin-3-one containing Δ4,5 and Δ2,3 double bonds [33].
Peptides
Cyclopeptides are cyclic molecules produced from either non-proteinogenic or proteinogenic amino acids linked by peptide or amine bonds, which are reported from fungi, plants, bacteria, mammals, sponges, and algae [39,40]. These metabolites exhibited various biological activities, including antibacterial, insecticidal, cytotoxic, and anticancer properties [39,40].
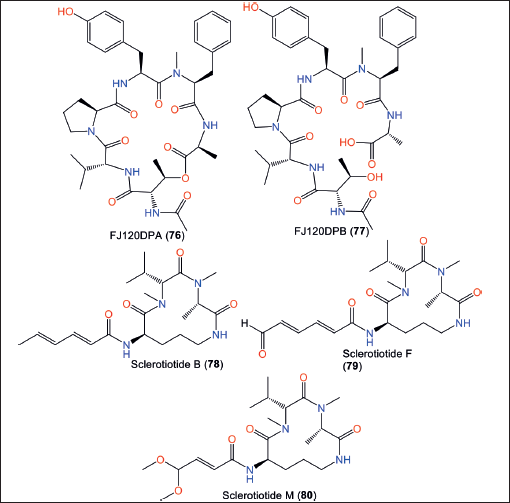
Cyclohexadepsipeptide 76 and its linear derivative 77, new metabolites were isolated from sea sediment-associated A. ochraceopetaliformis collected from the Jeju-do coast, Korea using SiO2/Sephadex LH-20 CC/HPLC [38]. Their structures were assigned using HR LC/MS-MS/spectral tools and Marfey’s method. Compound 76 has uncommon L-N-AcThr and D-Val units, whereas 77 was assigned as a N-AcThr and Ala ester linkage hydrolyzed derivative of 76 (Fig. 11). Compounds 76 and 77 had inhibition capacity for SrtA (IC50s 131.9 and 77.0 µM, respectively), compared to berberine chloride (IC50 104.3 µM) [38]; however, they were weakly active on isocitrate lyase (ICL). SrtA and ICL are prominent targets for developing anti-virulence medications that combat pathogenic microbes [38].
 | Figure 11. Chemical structures of peptides (76–80) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
A novel cyclic tripeptide, 80, in addition to 78 and 79 were isolated from the culture EtOAc extract of sea-water-derived A. ochraceopetaliformis using SiO2/Sephadex LH-20 CC/HPLC [37]. Compound 80 consists of Ala, Orn, and Val amino acid residues with (E)-4,4-dimethoxy-1-one-2-butenyl side chain. Its 2S/6S/12S configuration was assured based on calculated nuclear magnetic resonance chemical shifts coupled with DP4+ statistical method and optical rotation. Compounds 78 and 79 had selective anti-fungal properties against Candida albicans with (MICs 3.8 and 30 µM, respectively). On the other hand, 78–80 revealed weak cytotoxicity against BXPC-3 and HPAC cell lines in the CCK-8 assay [37].
Polyketides
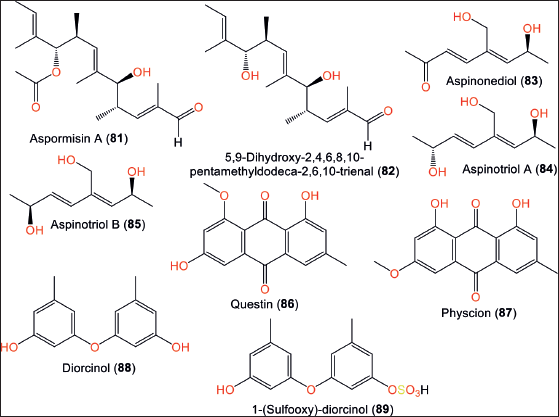
Aspormisin A (81) a new linear polyketide and its related metabolite, 82 were obtained from SCSIO-41020 derived from Hypnea pannosa alga collected from the South China Sea/Hainan province/China. Compound 81 has a C-9 acetate group instead of C-9 OH in 82 (Fig. 12; Table 5).
 | Figure 12. Chemical structures of polyketides (81–85), anthraquinones (86 and 87), and biphenyl ethers (88 and 89) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
 | Table 5. List of polyketides, anthraquinones, diphenyl ethers, and other metabolites isolated from Aspergillus ochraceopetaliformis. [Click here to view] |
Compound 82 demonstrated a notable NO production inhibitory potential, compared to 81, suggesting that the C-9 OH in 82 has a substantial role in the activity. Additionally, 82 effectively prohibited the in vitro and in vivo release of pro-inflammatory cytokines (IL-6, MCP-1, and TNF-α) caused by LPS [29]. Hu et al. [11] reported the isolation of pentaketides 83–85, which were formerly separated from Aspergillus ostianus 01F313 cultured with bromine-modified artificial seawater [41].
Anthraquinones and biphenyl ethers
The anthraquinones, 86 and 87 reported by [10] had weak capacities versus different Gram-positive and -negative bacteria and fungi strains [10]. Park et al. [34] reported biphenyl ethers: 88 and its new sulfonated derivative 89 from marine sponge-derived strain. Compound 88 was reported from other Aspergillus species such as Aspergillus nidulans, Aspergillus versicolor, and Aspergillus tennesseensis. These compounds displayed no antimicrobial and SrtA and ICL inhibitory activity [34].
Other metabolites
Compounds 90–92 were examined for antimicrobial and cytotoxic activities. Compound 92 displayed cytotoxic potential versus B16 (IC50 1.0 µM) in the MTT assay, whereas 91 exhibited weak antimicrobial activity against Bacillus subtilis, Micrococcus luteus, and Staphylococcus aureus (MICs 128 µg/ml) in the microporous plate method [30]. The biphenyl glycoside: 93 and triterpene 95 exhibited powerful antifungal and antibacterial properties (MICs 0.09–0.87 mg/ml) in the agar dilution technique [10] (Fig. 13). From the SCSIO-05702 strain, 94 and 97 were identified [12]. Additionally, 96 was obtained from glycyrrhetinic acid biotransformation using A. ochraceopetaliformis ATCC-12066 which has a C-2 and C-3 double bond, instead of the 3-OH group in glycyrrhetinic acid [42].
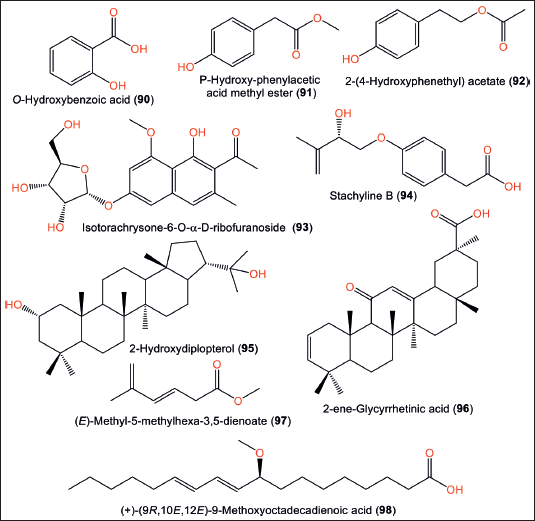 | Figure 13. Chemical structures of other metabolites (90–98) reported from Aspergillus ochraceopetaliformis. [Click here to view] |
BIOLOGICAL ACTIVITIES OF A. OCHRACEOPETALIFORMIS EXTRACTS
Abd El-Rahman et al. reported that EtOAc extract of MN083316 strain obtained from coral reefs collected from Ein El-Sukhna/Red Sea/Egypt exhibited powerful cytotoxic potential versus Hep-G2 cell line (C50 18.8 µg/ml) [36]. A study by Ramdass and Rampersad [43] revealed that A. ochraceopetaliformis isolated from Marac–Moruga mud volcano/South Trinidad possessed the broadest spectrum of oxidation against different tested substrates that completely decolorized methylene blue after 20 days. It was shown to be one of the strains with the highest oil tolerance and biosurfactant production [43]. Pattnaik et al. investigated the impact of A. ochraceopetaliformis SSP13 on reducing Pseudomonas aeruginosa PAO1’s QS-regulated virulence factors and biofilm production. The extract was found to prohibit the synthesis of pyocyanin, exopolysaccharides, rhamnolipids, LasA protease, LasB elastase, and chitinase, as well as causing notable changes in P. aeruginosa PAO1 motility and biofilm development. In the gas chromatography-mass spectrometry (GC-MS) analysis, 3,5-di-tert butyl phenol, benzaldehyde, 4-ethoxy, 3-benzofuran carboxaldehyde, 2-methoxy, furan, 2,2′-methylenebis[5-methyl], and vermelone were the main bioactive components identified by GC-MS analysis that were linked to the extract quorum sensing attenuating potential by targeting some receptor proteins [44].
DISCUSSION AND CONCLUSION
Aspergillus ochraceopetaliformis is a rich source of bioactive compounds with significant chemical diversity. The current review provides a thorough overview of A. ochraceopetaliformis, with a varied secondary metabolite profile. This review reveals that the Pyran derivatives represent the most abundant (24 compounds) metabolites, followed by alkaloids (16 compounds) and other classes such as cyclopentanones, lactones, sesquiterpenoids, and isocoumarins (Fig. 14). These metabolites were abundant through diverse environmental habitats, including soil, sponges, endophytes, algae, sea sediment, and seawater. The major number of metabolites were reported from soil-, sponges-, and endophytes-associated strains (Fig. 15).
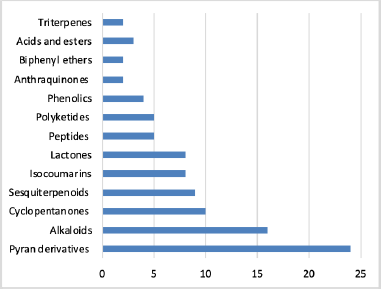 | Figure 14. Different classes of compounds reported from Aspergillus ochraceopetaliformis. [Click here to view] |
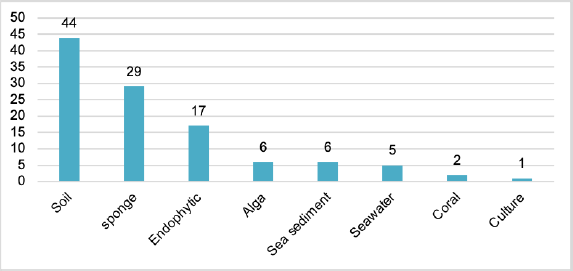 | Figure 15. Number of metabolites reported from Aspergillus ochraceopetaliformis isolated from different sources. [Click here to view] |
Multiple studies have shown that the production of A. ochraceopetaliformis metabolites majorly driven by biosynthetic gene clusters (BGCs), which encode enzymes such as non-ribosomal peptide synthetases (NRPSs), polyketide synthases, and hybrid pathways [45,46]. Genomic analyses have identified a broad array of BGCs responsible for synthesizing bioactive compounds like diketopiperazines, ochratoxins, and meroterpenoids [47–49]. Recent epigenetic and transcriptomic research further indicates that gene expression within these clusters is regulated by DNA methylation and histone modifications, ultimately influencing metabolite production [50]. Further characterization of these biosynthetic pathways could offer valuable insights for metabolic engineering approaches aimed at producing biologically active high-value compounds.
Among the identified metabolites, lactones 36, 37, and 39 have shown inhibitory effects on superoxide anion production and elastase release. However, their precise molecular targets remain unknown, limiting our understanding of their anti-inflammatory mechanisms. Additionally, variability in culture conditions across studies poses a challenge for comparative analysis. For example, sesquiterpenoids such as ochracenes A–I were only produced in agitated, nutrient-rich fermentation environments, whereas pyran derivatives were obtained under nutrient-deprived conditions—demonstrating the strong impact of environmental factors on metabolite biosynthesis.
Future research should integrate omics-based profiling, molecular docking, and gene knockout techniques to identify specific targets and enhance the efficiency of metabolite production. Additionally, advancements in genetic engineering—particularly CRISPR/Cas-based pathway modifications—could significantly boost the production of valuable metabolites like alkaloids and diketopiperazines, increasing their pharmaceutical potential. Addressing these knowledge gaps will improve our understanding of the metabolites produced by A. ochraceopetaliformis and their therapeutic applications. In conclusion, A. ochraceopetaliformis is a promising source of bioactive compounds with notable biological activities, and further investigation may uncover new treatments for a variety of diseases.
LIST OF ABBREVIATIONS
A-549, human lung adenocarcinoma epithelial cell line; B16, mouse melanoma cell line; BEL-7042, human hepatocellular carcinoma cell line; BXPC-3, human pancreas adenocarcinoma cell line; CCK-8, cell counting kit-8; DU145, human prostate carcinoma cell line; ECD, electronic circular dichroism; EtOAc, ethyl acetate; EV, extracellular vesicles; fMLP, N-formyl-L-methionyl-L-leucyl-L-phenylalanine; GC-MS, gas chromatography mass spectrometry; HepG2, human hepatocellular liver carcinoma cell line; HeLa, human cervical epitheloid carcinoma cell line; HL-60, human promyelocytic leukemia cell line; HMEC-1, human microvascular endothelial cell line; HPAC, human pancreatic adenocarcinoma cell line; HPLC, high-performance liquid chromatography; HT-29, human colon cancer cell line; ICL, isocitrate lyase; IL-6, interleukin-6; K-562, human erythroleukemic cell line; LPS, lipopolysaccharide; MCF-7, human breast adenocarcinoma cell line; MCP-1, monocyte chemoattractant protein-1; MDA-MB-231, human breast cancer cell line; MIC, minimum inhibitory concentrations; MTT, 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; MVBs, multivesicular bodies; NO, nitric oxide; NOESY, nuclear overhauser effect spectroscopy; RP-18 CC, reversed phase-18 column chromatography; SGC-7901, human gastric adenocarcinoma cell line; SrtA, sortase A; THP-1, human monocytic cell line; TNF-α, tumor necrosis factor alpha; U937, pro-monocytic, human myeloid leukaemia cell line.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
This study is supported via funding from Prince Sattam bin Abdulaziz University project number (PSAU/2025/R/1446).
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
The authors confirm that the data supporting the findings of this study are available within the article.
CONSENT FOR PUBLICATION
All authors have read and approved the final manuscript.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Hawksworth DL, Lücking R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr. 2017;5(4):1-17. CrossRef
2. Blackwell M. The fungi: 1, 2, 3… 5.1 million species? Am J Bot. 2011;98:426–38.
3. Gori K, Cantor MD, Jakobsen M, Jespersent L, Nout M, Aidoo KE, et al. Traditional food and beverage fermentation. In: Esser K, Hofrichter M, editors. The mycota, industrial applications. 2nd ed. Berlin, Germany: Springer; 2010. pp. 3–97.
4. Demain AL, Velasco J, Adrio JL. Industrial mycology: past, present, and future. In J. W. Bennett, Paul A. Lemke, editors. Handbook of industrial mycology. Boca Raton, FL: CRC Press; 2004. pp. 20–45.
5. Alam S, Nisa S, Daud S. Mycotoxins in environment and its health implications. In: Ahmed T, Hashmi MZ, editors. Hazardous environmental micro-pollutants, health impacts and allied treatment technologies. Emerging contaminants and associated treatment technologies. Cham, Switzerland: Springer; 2022. pp. 289–318. CrossRef
6. Hussein MA, Gherbawy YA, Abd El-sadek MS, Al-Harthi HF, GAM El-Dawy E. Phylogeny of Aspergillus section Circumdati and inhibition of ochratoxins potential by green synthesised ZnO nanoparticles. Mycology 2024;1–12. CrossRef
7. Abdel-Azeem AM, Salem FM, Abdel-Azeem MA, Nafady NA, Mohesien MT, Soliman EA. Chapter 1 - Biodiversity of the genus Aspergillus in different habitats. In: Gupta VK, editor. New and future developments in microbial biotechnology and bioengineering. Amsterdam, Netherlands: Elsevier; 2016. pp: 3–28. CrossRef
8. Wang J, Wei X, Qin X, Tian X, Liao L, Li K, et al. Antiviral merosesquiterpenoids produced by the antarctic fungus Aspergillus ochraceopetaliformis SCSIO 05702. J Nat Prod. 2016;79:59–65.
9. Liu J, Wu W, Cao M, Yang F, Lin H. Trienic α-pyrone and ochratoxin derivatives from a sponge-derived fungus Aspergillus ochraceopetaliformis. Nat Prod Res. 2018;32:1791–7.
10. Asmaey MA, Abatis D, Abdel-Razek AS, Lambrinidis G, Chinou I, Fokialakis N, et al. Ochraceopyronide, a rare α-pyrone-C-lyxofuranoside from a soil-derived fungus Aspergillus ochraceopetaliformis. Molecules 2021;26:3976.
11. Hu H, Li C, Tsai Y, Yang D, Wu Y, Hwang T, et al. Secondary metabolites and bioactivities of Aspergillus ochraceopetaliformis isolated from Anthurium brownii. ACS Omega 2020;5:20991–9.
12. Wang J, He W, Kong F, Tian X, Wang P, Zhou X, et al Ochracenes A–I, humulane-derived sesquiterpenoids from the Antarctic fungus Aspergillus ochraceopetaliformis. J Nat Prod. 2017;80:1725–33.
13. Guo C, Wang P, Pang X, Lin X, Liao S, Yang B, et al. Discovery of a dimeric zinc complex and five cyclopentenone derivatives from the sponge-associated fungus Aspergillus ochraceopetaliformis. ACS Omega 2021;6:8942–9.
14. Ghazawi KF, Fatani SA, Mohamed SG, Mohamed GA, Ibrahim SR. Aspergillus nidulans—natural metabolites powerhouse: structures, biosynthesis, bioactivities, and biotechnological potential. Fermentation 2023;9:325.
15. Hareeri RH, Aldurdunji MM, Abdallah HM, Alqarni AA, Mohamed SG, Mohamed GA, et al. Aspergillus ochraceus: metabolites, bioactivities, biosynthesis, and biotechnological potential. Molecules 2022;27:6759.
16. El-Agamy DS, Ibrahim SR, Ahmed N, Khoshhal S, Abo-Haded HM, Elkablawy MA, et al. Aspernolide F, as a new cardioprotective butyrolactone against doxorubicin-induced cardiotoxicity. Int Immunopharmacol. 2019;72:429–36.
17. Ibrahim SR, Elkhayat ES, Mohamed GA, Khedr AI, Fouad MA, Kotb MH, et al. Aspernolides F and G, new butyrolactones from the endophytic fungus Aspergillus terreus. Phytochem Lett. 2015;14:84–90.
18. Ibrahim SR, Pinzari F, Mohamed SG, Hussein HG, Alzain AA, Almogaddam MA, et al. Compilation of secondary metabolites produced by fungal strains identified as Aspergillus flavus var. oryzae (formerly Aspergillus oryzae), with a review of biotechnological applications, and computational studies of their anti-COVID-19 activity. Mycol Prog. 2024;23:1–47.
19. Ibrahim SR, Abdallah HM, Mohamed GA, Deshmukh SK. Exploring potential of Aspergillus sclerotiorum: secondary metabolites and biotechnological relevance. Mycol Prog. 2023;22:1–28.
20. Mohamed GA, Ibrahim SR, El-Agamy DS, Elsaed WM, Sirwi A, Asfour HZ, et al. Terretonin as a new protective agent against sepsis-induced acute lung injury: impact on SIRT1/Nrf2/NF-κBp65/NLRP3 signaling. Biology 2021;10:1219.
21. Ibrahim SR, Mohamed GA, Kamal HM, Mohamed SG, Khedr AI. Terretonins from Aspergillus genus: structures, biosynthesis, bioactivities, and structural elucidation. Mini-Rev Org Chem 2022;19:257–69.
22. Ibrahim SR, Mohamed GA, Khedr AI. γ-Butyrolactones from Aspergillus species: structures, biosynthesis, and biological activities. Nat Prod Commun 2017;12:791–800.
23. Alberts AW. Lovastatin and simvastatin-inhibitors of HMG CoA reductase and cholesterol biosynthesis. Cardiology 1990;77:14–21.
24. Visagie CM, Varga J, Houbraken J, Meijer M, Kocsubé S, Yilmaz N, et al. Ochratoxin production and taxonomy of the yellow aspergilli (Aspergillus section Circumdati). Stud Mycol. 2014;78:1–61.
25. Brasch J, Varga J, Jensen J, Egberts F, Tintelnot K. Nail infection by Aspergillus ochraceopetaliformis. Med Mycol. 2009;47:658–62.
26. Akter H, Ara I, Alam NB, Alam N. The first report of Aspergillus ochraceopetaliformis Bat. & Maia from small indigenous dry fish Setipinna phasa (Hamilton 1822). Jahangirnagar Univ J Biol Sci. 2023;12(1):87–95.
27. Ngo CC, Nguyen QH, Nguyen TH, Quach NT, Dudhagara P, Vu THN, et al. Identification of fungal community associated with deterioration of optical observation instruments of museums in Northern Vietnam. Appl Sci. 2021;11:5351.
28. Dai Q, Zhang F, Feng T. Sesquiterpenoids specially produced by fungi: structures, biological activities, chemical and biosynthesis (2015–2020). J Fungi 2021;7:1026.
29. Chen C, Ren X, Tao H, Cai W, Chen Y, Luo X, et al. Anti-inflammatory polyketides from an alga-derived fungus Aspergillus ochraceopetaliformis SCSIO 41020. Mar Drugs 2022;20:295.
30. Zhong R, Shi C, Wu X, Zou Y, Li J, Liang Y. A new isocoumarin from sponge endophytic fungus Aspergillus ochraceopetaliformis with cytotoxic activity. Nat Prod Res. 2024;1–6.
31. Mitani F, Lin J, Sakamoto T, Uehara R, Hikita T, Yoshida T, et al. Asteltoxin inhibits extracellular vesicle production through AMPK/mTOR-mediated activation of lysosome function. Sci Rep. 2022;12:6674.
32. Cong M, Ren X, Song Y, Pang X, Tian X, Liu Y, et al. Ochrathinols A and B, two pairs of sulfur-containing racemates from an Antarctic fungus Aspergillus ochraceopetaliformis SCSIO 05702 inhibit LPS-induced pro-inflammatory cytokines and NO production. Phytochemistry 2023;208:113593.
33. Jiao S, Zhang R, Li J, Wuken S, Zhang HX, Tu P, et al. Phytochemical and pharmacological progress on humulane-type sesquiterpenoids. Zhongguo Zhong Yao Za Zhi. 2018:43(22):4380–90.
34. Park SC, Lee J, Hwang J, Kwon O, Liao L, Oh D, et al. Ochraceopetalin, a mixed-biogenetic salt of polyketide and amino acid origins from a marine-derived Aspergillus ochraceopetaliformis fungus. Mar Drugs 2021;19:413.
35. Asmaey M, Abatis D, Abdel-Razek A, Chinou I, Fokialakis N, Shaaban M, et al. New indole diketopiperazine alkaloids from a soil-derived fungus Aspergillus ochraceopetaliformis. Planta Med. 2021;87:PC9–70.
36. Abd El-Rahman TM, Tharwat NA, Abo El-Souad SM, El-Beih AA, El-Diwany AI. Biological activities and variation of symbiotic fungi isolated from Coral reefs collected from Red Sea in Egypt. Mycology 2020;11:243–55.
37. Liao Y, Hong X, Yang J, Qin J, Zhang B, Lin J, et al. Structure elucidation of a novel cyclic tripeptide from the marine-derived fungus Aspergillus ochraceopetaliformis DSW-2. Nat Prod Res. 2022;36:3572–8.
38. Hwang J, Lee J, Park SC, Lee J, Oh D, Oh K, et al. New peptides from the marine-derived fungi Aspergillus allahabadii and Aspergillus ochraceopetaliformis. Marine Drugs 2019;17:488.
39. Gang D, Kim DW, Park H. Cyclic peptides: promising scaffolds for biopharmaceuticals. Genes 2018;9:557.
40. Lee Y, Phat C, Hong S. Structural diversity of marine cyclic peptides and their molecular mechanisms for anticancer, antibacterial, antifungal, and other clinical applications. Peptides 2017;95:94–105.
41. Kito K, Ookura R, Yoshida S, Namikoshi M, Ooi T, Kusumi T. Pentaketides relating to aspinonene and dihydroaspyrone from a marine-derived fungus, Aspergillus ostianus. J Nat Prod. 2007;70:2022–5.
42. Xie Z, Gu X, Dekebo A, Wei S, Hu X. Two new compounds from biotransformation of glycyrrhetinic acid. Nat Prod Res. 2024;1–5. CrossRef
43. Ramdass AC, Rampersad SN. Biodiversity and biocatalyst activity of culturable hydrocarbonoclastic fungi isolated from Marac–Moruga mud volcano in South Trinidad. Sci Rep. 2021;11:19466.
44. Pattnaik S, Ahmed T, Ranganathan SK, Ampasala DR, Sarma VV, Busi S. Aspergillus ochraceopetaliformis SSP13 modulates quorum sensing regulated virulence and biofilm formation in Pseudomonas aeruginosa PAO1. Biofouling 2018;34:410–25.
45. Gluck-Thaler E, Haridas S, Binder M, Grigoriev IV, Crous PW, Spatafora JW, et al. The architecture of metabolism maximizes biosynthetic diversity in the largest class of fungi. Mol Biol Evol. 2020;37:2838–56.
46. Zhang S, Shi G, Xu X, Guo X, Li S, Li Z, et al. Global analysis of natural products biosynthetic diversity encoded in fungal genomes. J Fungi. 2024;10:653.
47. Dai G, Shen Q, Zhang Y, Bian X. Biosynthesis of fungal natural products involving two separate pathway crosstalk. J Fungi. 2022;8:320.
48. Ferrara M, Gallo A, Perrone G, Magistà D, Baker SE. Comparative genomic analysis of ochratoxin A biosynthetic cluster in producing fungi: new evidence of a cyclase gene involvement. Front Microbiol. 2020;11.
49. Wei B, Ying T, Lv H, Zhou Z, Cai H, Hu G, et al. Global analysis of fungal biosynthetic gene clusters reveals the diversification of diketopiperazine biosynthesis. Bioresour Technol. 2025;132218.
50. Das S, Kwon M, Kim JY. Enhancement of specialized metabolites using CRISPR/Cas gene editing technology in medicinal plants. Front Plant Sci. 2024;15:1279738.