INTRODUCTION
Cartilage tissue engineering, particularly with advancements in stem cell engineering, plays a crucial role in achieving the United Nations Sustainable Development Goal 3: Good Health and Well-being, which aims to promote healthy lives for people of all ages. Due to the limited self-repair capacity of cartilage, conditions such as osteoarthritis and traumatic cartilage injuries pose significant healthcare challenges worldwide, especially among aging populations. The development of tissue engineering that integrates biomaterials and stem cell technologies aims to create innovative regenerative solutions for cartilage-related diseases, which affect millions globally and often lead to long-term disability. By offering effective, minimally invasive, and patient-specific therapies, cartilage tissue engineering contributes to sustainable healthcare, improving the quality of life while reducing the burden on healthcare systems.
Human Wharton’s jelly-derived mesenchymal stem cells (hWJ-MSCs) are one cell source frequently used in cartilage tissue engineering and tissue regeneration development [1]. The role of the microenvironment significantly impacts how hWJ-MSCs cells display their phenotype, particularly when an extracellular matrix (ECM) is present to act as a substrate for controlling the stem cell differentiation process [2]. ECM is a cell microenvironment that not only functions as a provider of biophysical signals but also as a biochemical signal that directs hWJ-MSCs differentiation into chondrocyte. Stimulus from engineering and modification of topography influenced hWJ-MSCs ECM remodeling to induce the differentiation process [3]. The development of nanotopographic engineering technologies to regulate cell behavior and differentiation may be a solution for gaining knowledge of the interactions and signaling pathways that occur throughout mesenchymal stem cell regulation and differentiation [4, 5].
The microenvironment of cells in the body has nanoscale features; even nanotopography has been found everywhere, especially in the ECM around cells [6]. As nanotechnology continues to advance, nanopatterned substrates can be increasingly integrated. The extent to which nanotechnology influences the creation of a cell microenvironment will have a significant impact on the differentiation and behavior of cells, which are dependent on the properties of the substrate [7]. Research conducted by Kim et al. [8] showed that the expression of osteogenesis markers such as COL1, RUNX2, and OPN was higher in cells cultured on nanopatterned substrate surfaces. Another study showed that in vitro study, improved cell alignment is observed in an ECM microenvironment that is integrated into linear geometries by introducing grooves and grids into the substrate surface. The dimensions of the grooves and protrusions with a width of 35 nm-25 mm and a depth of 14 nm-5 mm can induce good cell alignment [9].
Nanolithography techniques, such as nanoimprint lithography, microscope nanolithography, electron beam lithography, and focused ion beam lithography, are currently being explored for their high-precision capabilities in generating minute surface patterns, referred to as nanopatterning techniques [8,10]. However, some of these methods are not the preferred options due to the use of solvents and materials that are hazardous and toxic to cell viability. Additionally, the mass production of substrates with nanoscale patterns is constrained by the complexity, high cost, low efficiency, and time-consuming nature of the manufacturing processes [10]. Fabrication of nanotopographic substrates using the above methods is complex and requires routine integration into cell cultures. To overcome this problem, the existence of commercially available optical discs such as Bluray-disc Recordable (BD-R) is an advantage because it combines a nanopatterned polycarbonate (PC) substrate with many concentric ring-shaped nano grooves that can be used for cell culture. Therefore, such optical discs can be used directly as templates for printing substrate patterns on cell cultures after appropriate pre-processing of the discs.
In this research, spidroin can be applied to support the performance of the substrate as a place to grow cell cultures. One kind of spider frequently found in Indonesia is the Argiope family spider [11]. It is known that proteins from this species spider silk promote hWJ-MSC cell attachment and chondrogenic development [12]. These spiders weave a web called silk spidroin, which may nowadays be used as a biomaterial for scaffolding. Because silk spidroin possesses the strength, porosity, biodegradability, and biocompatibility required for cell proliferation and differentiation, it can be utilized as a scaffold material for cartilage tissue engineering [13,14].
A naturally occurring substance called silk spidroin is reported to contain the amino acid sequence Arginine-Glycine-Aspartic Acid (a peptide sequence) (RGD) (tripeptide Arg-Gly-Asp) [15,16]. Numerous interactions exist between the surrounding matrix and the cells through this RGD sequence. Similar to fibronectin, which also has the RGD sequence, this sequence binds onto integrins, which are cell transmembrane proteins [17]. The attachment, migration, and proliferation of hWJ-MSC cells can be enhanced by the RGD sequence present in the silk spidroin of the Argiope appensa spider [16]. Consequently, by guiding the chondrogenic development of hWJ-MSCs cells, the combination of polydimethylsiloxane (PDMS)-BD-R nanopatterns and spider web extract (spidroin) may offer a novel approach to creating improved cartilage tissue engineering method.
MATERIALS AND METHODS
The overall methodology of this study consists of three main procedures: isolation and characterization of human Mesenchymal Stem Cells (hMSCs), fabrication of nanopatterned-based Bluray discs and extraction of spidroin protein from raw spider webs. A comprehensive illustration of the entire workflow is presented in Figure 1.
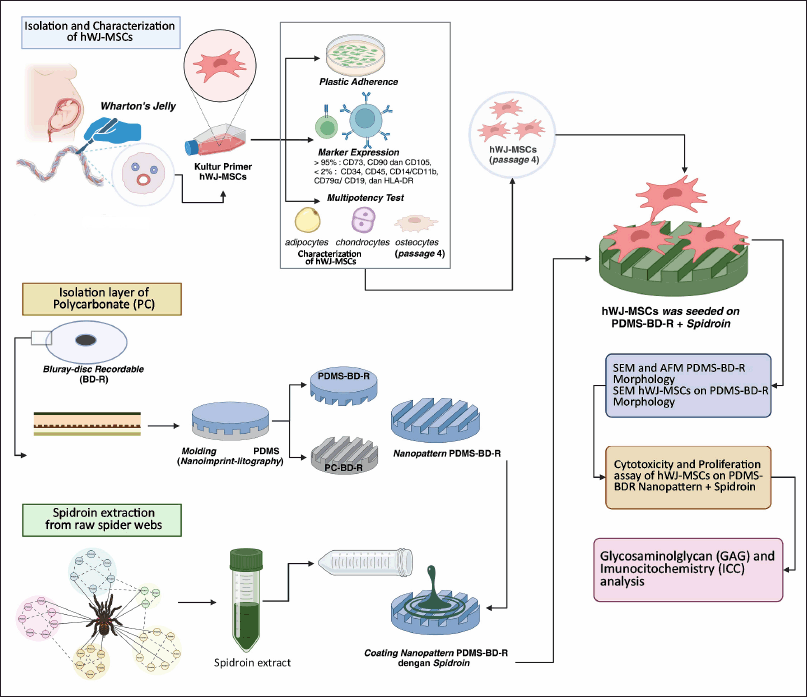 | Figure 1. An illustration of schematic research design. [Click here to view] |
Isolation and primary culture of human Wharton’s Jelly (hWJ)
The primary culture method used is the Explant Method [18]. The hWJ samples from the umbilical cord were collected at the Rumah Sakit Umum Daerah Kiwari Kota Bandung with proper informed consent and ethical approval (SK. 316/UN6.KEP/EC/2023). The donor’s placental tissue is put in a sterile container with a transport medium of sterile Phosphate-buffered saline (PBS) and 1% antibiotic-antimycotic (ABAM) solution. The samples were then cleaned with 70% alcohol for 30 seconds, 10% povidone-iodine for 30 seconds, and thrice rinses with sterile PBS containing 4% ABAM. The hWJ tissue was divided longitudinally, and the epithelium and blood vessels were removed. The tissue is then incised and sliced into 3–5 mm pieces with a sterile knife linked to a scalpel. After that, the tissue was put in a T-25 flask and kept at room temperature until it was attached. In primary culture, the growth media consists of Dulbecco-modified eagle medium (DMEM, Gibco), 10% fetal bovine serum (FBS, Gibco), and 1% ABAM (Gibco) gently given to the tissue cultured in a flask, T-25. The flask was then incubated at 37°C, 5% CO2, and the medium changed every 2–3 days. After achieving more than 80% confluence, cells were collected using Trypsin-EDTA 0.25% (Gibco).
Analysis of cell surface antigen expression
The sample consisted of 5 × 105 cells at passage 4% and 0.25% trypsin or Ethylenediaminetetraacetic acid was used to remove cultured cells from the T-25 flask substrate. The phenotypic of hWJ-MSCs was assessed by fluorescence-activated cell sorting (FACS) following the Human MSC Analysis Kit procedure (BD Stemflow protocol) and analyzed using a flow cytometer (BD FACS Aria III) and BD FACS Version 6.1.3TM software.
Multipotency test analysis of differentiation of hWJ-MSCs
The hWJ-MSCs multidifferentiation experiment assessed the cells’ ability to develop into chondrocytes, adipocytes, and osteocytes. Cells with density 2 × 104 (passage 4/5) were grown on 24 well plates with chondrogenic, adipogenic, and osteogenic differentiation medium (Stepro differentiation media; Gibco). The cells were then incubated at 37°C, 5% CO2, and the media changed every 2–3 days. After 21 days, the cells were preserved with 4% formaldehyde solution. Cell differentiation was investigated by staining cells with Alcian Blue as chondrocytes, Alizarin Red for osteocytes, and Oil Red O for adipocytes. The staining results were analyzed using an optical phase contrast microscope.
Viscosity, zeta potential, and Raman spectroscopy analysis
The viscosity and zeta potential of the spidroin solution were then determined using a Horiba Scientific Nanoparticle Analyzer SZ using a dynamic light scattering technique. A syringe transferred 1 ml of spidroin solution to the cuvette. The protocol for extracting spidroin from raw spider silk can be found in the supplementary material. All measurements were carried out in triplicate at 25°C. Results are presented as mean ± standard deviation (SD) (n = 3). Spider webs were characterized using Raman Spectroscopy with the HORIBA Micro Raman Standard Microscopy Spectroscopy instrument with a wavelength of 532 nm. Results are reported as mean ± SD (n = 3).
Cytotoxicity test of silk spidroin extract
The concentration of spider web extract solution was evaluated using the Bradford technique, with concentrations tested at 25, 50, 100, 250, 500, and 1,000 µg/ml. A 96-well plate was coated with 100 µl of spidroin extract solution in DMEM medium to conduct the test. The plate was then incubated for 1 hour at 37°C with 5% CO2. The (3 [4,5 - dymethylthiazol-2y1] - 2,5 - dyphenylthiazolium bromide) (MTT) technique was used to perform the cytotoxicity test. hWJ-MSC cells at a density of 5 × 103 cells were sown in 96-well plates and cultured in an incubator at 37°C and 5% CO2 for 72 hours. After 72 hours, the culture medium was removed, and MTT reagent (0.5 mg/ml) was added to the DMEM LG media (no FBS). The plate was covered with aluminium foil (dark condition) and incubated for 4 hours at 37°C with 5% CO2. After incubation, purple formazan crystals are going to form on the plate. The remaining MTT reagent was carefully removed, the residual crystals were dissolved with 100 µl of sterile Dimethyl sulfoxide, and the absorbance was measured using a microplate reader at 595 nm.
PDMS-BD-R nanopattern fabrication
The protocol for fabricated BD-R based nanopattern could be found in the supplementary material.
Nanopattern morphology using scanning electron microscopy (SEM) and atomic force microscopy (AFM)
The PDMS-BD-R nanopattern substrate, (including nanopatterned and unpatterned), was characterized using SEM coated with gold with a sputter coater (MC100 Ion Sputter Coater, Hitachi Japan), and the SEM results were analyzed with ImageJ. Both of the PDMS-BD-R nanopattern substrates were characterized using AFM coated with gold with a sputter coater (Park System XE-70) and the AFM results were analyzed with ImageJ.
Plasma treatment and water contact angel (WCA) test
The PDMS-BD-R nanopattern substrate measuring 13 mm was placed in a sterile petri dish and inserted into the plasma treatment unit under low-pressure conditions for 2 minutes. Contact angle analysis is carried out immediately after plasma treatment to avoid stability problems due to storage. The contact angle was measured by analyzing the shape of water droplets on the PDMS surface. A total of 25 µl ddH2O was dropped perpendicularly on the surface of the PDMS film, and images were taken using a DinoLITE microscope camera and analyzed using DinoCapture2 software. The WCA is measured by taking the impact of the drop, the intersection point between the contact drop and the projected surface. If the contact angle is more than 80°, the surface is considered hydrophobic, and if it is more than 150°, it is considered super hydrophobic.
Seeding hWJ-MSC culture on PDMS-BD-R nanopattern
2 × 104 cells were grown on a 13 mm diameter PDMS BD-R nanopattern substrate, coated or not with spidroin extract. The culture was carried out for a full 21 days using low glucose complete media consisting of 10% FBS and 1% ABAM in DMEM and kept in an incubator at 37°C and 5% CO2.
Analysis of morphology and proliferation of hWJ-MSCs on PDMS-BD-R nanopattern
Morphological analysis was carried out using SEM. 5 × 104 hWJ-MSCs cells were grown on sterile PDMS-BD-R nanopattern with a diameter of 13 mm and cultured under complete DMEM LG medium conditions incubated at 37°C, 5% CO2. To see the interaction of cells with the substrate, after 72 hours post-seeding, the culture medium was discarded, and the cells were rinsed with PBS 3×. For SEM analysis, cells were fixed with 100 µl 2.5% (v/v) GTA in 0.1 M Na-Cacodylate buffer for 2 hours. The cells were then rinsed with 0.1 M Na-Cacodylate buffer three times and rinsed using a serial alcohol solution (30%–100%, 5 minutes each) and dried by dipping the substrate and cells in a mixture of 100% alcohol solution and HMDS (hexamethyldisilazane) 1:1. The substrate was then dried at room temperature overnight and coated with gold with a sputter coater (MC100 Ion Sputter Coatter, Hitachi Japan). The SEM results were analyzed with ImageJ.
hWJ-MSC proliferation curve on PDMS-BD-R nanopattern
Proliferation tests were carried out on days 1, 3, 5, 7, and 14 after seeding using the MTT method, which refers to the MTT cytotoxic assay method.
Matrix glycosaminoglycan (GAG) abundance analysis
Cells cultivated on the PDMS-BD-R nanopattern on days 7, 14, and 21 were fixed before being aspirated into the culture media. All surfaces were washed twice with PBS before being fixed in (1:1) methanol : acetone (4°C) solution for 3 minutes. The substrate was moved to a 1% Alcian Blue solution in 3% acetic acid. After 30 minutes of incubation in Alcian Blue at room temperature, the surfaces of the PDMS-BD-R nanopattern were washed three times with 3% acetic acid for 2 minutes each. After washing in ddH2O for 2 minutes, the surface of the PDMS-BD-R nanopattern was dried for imaging or immersed in a 1% SDS solution for 30 minutes on a 200 rpm pallet shaker to remove the alcian blue stain. The absorbance of the dissolved solution was measured at a 605 nm wavelength. Absorbance measurements were carried out with a UV-VIS spectrophotometer using a disposable cuvette and 1 ml sample. 1% SDS solution was used as a blank in spectrophotometer measurements.
Immunocytochemistry (ICC) analysis (Collagen II and SOX9)
Cells grown on the nanopattern for 7, 14, and 21 days were fixed with series methanol-DMEM (50%, 70%, 80%, 90%, and 100%) at 20°C before being washed by PBS three times at room temperature. Following fixation, cells were permeabilized with PBS-T (0.05% Tween 20 in PBS) for 20 minutes at room temperature. Cells were inhibited from nonspecific binding antibodies with 3% bovine serum albumin (BSA in PBST) for 60 minutes. Primary antibodies to collagen type II (ab34712, Abcam) and SOX9 (ab3697, Abcam) were added to the samples and incubated overnight at 4°C. Samples were rinsed three times with PBS before incubating with a secondary antibody (1:200 goat anti-rabbit IgG HNL Alexa Fluor 488 ab150077, Abcam) in the dark for 2 hours. After that, 0.1% Rhodamine phalloidin (Invitrogen, AB235138) and 4′,6-diamidino-2-phenylindole (DAPI, Thermo Fisher) were applied for 45 and 15 minutes, respectively, before being washed three times with PBS. The stained cells were examined using a confocal microscope (Olympus FV 1200 Confocal Laser Scanning Microscope).
Statistical analysis
Data were gathered from at least three repetitions and reported as mean ± SD. The statistical analysis was carried out using GraphPad Prism 9 software. Two-way ANOVA was used to compare differences (p < 0.05) in cytotoxicity, proliferation, and differentiation (Alcian Blue) through post-hoc analysis performed via Tukey’s multiple comparison test.
RESULTS AND DISCUSSION
Characteristics and multipotency test of hWJ-MSCs cells
Under conventional culture conditions, the primary culture of hWJ-MSCs that had migrated from hWJ explants (Fig. 2A) adhered to the flask surface and showed a fibroblast-like shape. The formation of lipid droplets from hWJ-MSCs cells that had differentiated into adipocyte cells was confirmed using Oil Red O staining (Fig. 2B). Using Alcian Blue staining to analyze the presence of the proteoglycan matrix, or GAG (Fig. 2C), hWJ-MSCs can differentiate into chondrocyte cells, which are distinguished by the formation of chondrocyte cell nodules and the presence of the proteoglycan matrix (dark blue). Meanwhile, the accumulation of calcium mineralization with Alizarin Red staining (Fig. 2D) shows the successful differentiation of hWJ-MSCs into osteocytes. Positive results of the multipotency test are indicated by yellow arrows (Fig. 2B–D).
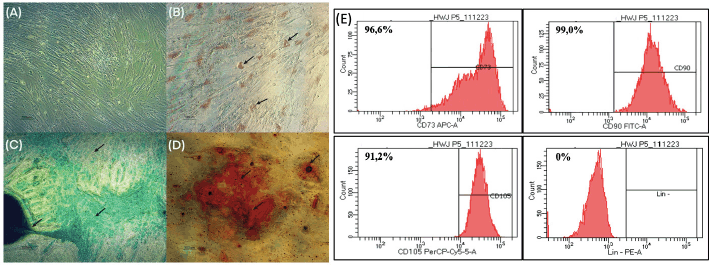 | Figure 2. Characterization of hWJ-MSC cultures. (A) Control group (no staining), (B) Oil Red O staining for adipocyte differentiation, (C) Alcian Blue staining indicating chondrogenic differentiation, and (D) Alizarin Red staining for osteocyte differentiation. The black arrow shows that each staining group’s marker had a favorable result. (E) CD73 APC-A positive marker (96.6%); CD90 FITC-A (99.0%) CD105 PerCP-Cy5-5-A (91.2%) and negative markers Lin–PE-A (CD34, CD45, CD11B, CD19, and HLA-DR PE (0%). [Click here to view] |
The results of FACS analysis (Fig. 2E) showed the presence of positive markers CD90 (96.6%), CD73 (99%), and CD105 (91.2%). The negative marker (Lin-negative) was obtained at 0%. In this study, the percentage of positive markers for CD90 and CD73 met the International Society for Cellular Therapy (ISCT) requirements with a rate of >95%, while CD105 was still <95%. Even though the CD105 percentage is below the value required by ISCT, it does not affect its multipotency ability in this study. This is consistent with studies by Pham et al. [19] that show the diverse capabilities of MSCs derived from the umbilical cord, especially when the cells are heterogeneous. The difference in expression of CD105(+)-hMSCs and CD105(-)-hMSCs reflects differences in immune system modulation abilities but does not affect their potential to differentiate in vitro into adipocyte, chondrocyte, and osteocyte cell lines. Variations in CD105 percentage values also depend on differences in hWJ-MSCs culture conditions, isolation methods, cell sources, and length of culture time. One of the influencing culture conditions is being over-confluent before the FACS examination, causing lower CD105 values [19,20]. The multipotency test and FACS characterization data indicate that the hWJ-MSCs cell culture fulfils the ICST requirements described in Dominici et al. [21].
Characterization of the spidroin A. appensa
Zeta potential and viscosity
The physical characteristics of the spidroin extract were analyzed based on the stability of the colloidal particles and their flow properties through zeta potential and viscosity values. Zeta potential can be defined as a parameter that shows the repulsion between charged particles in a colloid or suspension dispersion system [22]. The zeta potential value of a colloidal dispersion system to maintain the stability of its nano dispersion ranges below −30 mV or above +30 mV [22,23]. In Figure 3 below, the spidroin zeta potential value is obtained −34.56 mV, which indicates that this value is in the range required for a dispersion system to maintain its physical stability. Apart from representing the stability of the dispersion system, the optimum zeta potential value also determines the main factor in the initial adsorption of particles on the cell membrane [22,24], such as the interaction between spidroin proteins and hWJ-MSCs cells when the cells perform focal adhesion (FA). Meanwhile, viscosity is defined as the system’s resistance to gradual deformation of a medium, which is related to differences in strain rates in the flowing medium [25]. In this study, the spidroin solution extract had a viscosity value of 0.896 mPa.s, which is close to the viscosity value of water (0.89 mPa.s), so it shows good rheological properties because the solution flows relatively easily.
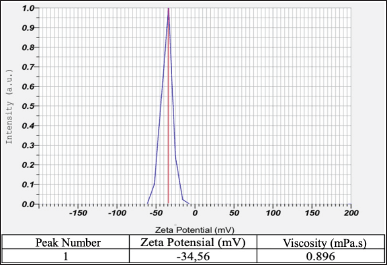 | Figure 3. Characterization of the spidroin A. appensa. The viscosity and zeta potential values of spidroin show a range of values indicating good and relatively stable physicochemical properties. [Click here to view] |
Raman spectroscopy analysis
Figure 4A above shows the results of Raman spectroscopy of A. appensa spider webs, which were analyzed at a wavelength of 532 nm, and the wave peak was projected between wave numbers 500 to 2,000 cm−1 [26]. The pink box area (Fig. 4A) shows the possible distribution of RGD sequences in spider webs that fall between the wave number ranges 900–1,100 cm−1 and 1,250–1,525 cm−1. This area has a more dominant distribution of the amino acids, arginine, glycine, and aspartate acid compared to other areas, which are thought to be the amino acids that form the RGD sequence [27]. The possibility of an RGD sequence detected at this wave number is obtained based on the peak that appears in the form of a Raman shift, which is compared with available literature data that refers to supplementary information [28–30]. The possible content of amino acids and secondary structures that form proteins that appear between wave numbers 500 to 2,000 cm−1 could be found in Table S45 on supplementary material.
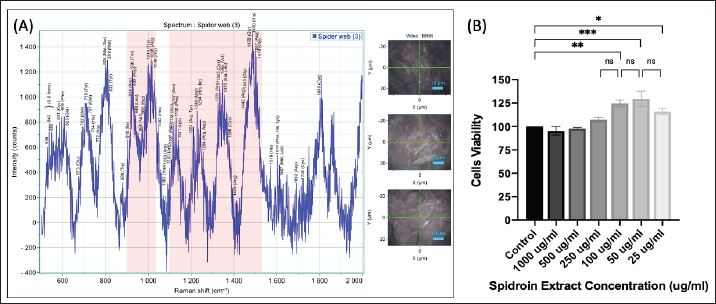 | Figure 4. Raman spectroscopy analysis of spider webs (Wavelength: 532 nm). (A) The area in the pink box represents the possible RGD sequences. (B) Cytotoxicity test of spider web extract on hWJ-MSC cells as measured by the MTT assay. Data are mean ± SD with significance markers * (p < 0.05), ** (p < 0.01), and *** (p < 0.001). [Click here to view] |
Spidroin cytotoxicity test
The MTT assay works by evaluating the metabolic activity of cells, where viable cells with active metabolism convert the MTT reagent into a purple formazan product. The amount of formazan produced correlates with the number of viable cells, allowing researchers to infer cell viability and indirectly assess cytotoxicity. The graph (Fig. 4B) describes the most optimal proliferation conditions for hWJ-MSCs cells at a spider web extract concentration of 50 and 100 µg/ml. Under these conditions, the percentage of hWJ-MSCs cell proliferation was above 125% compared to controls. The spidroin concentration range starts from 25 up to 1,000 µg/ml and produces cell viability >90%. This indicates that spider web extract is not toxic to cells in this concentration range. The concentration used in this study was 50 µg/ml. This determination refers to the standard testing protocol established by Altomare et al. [31] for the production of micropattern substrates coated with fibronectin protein.
Characterization of PDMS BR-D nanopatterns
SEM confirmed the successful completion of the fabrication process. Figure 5A–C below shows the SEM results of the nanopattern surface found on the PC-BD-R molding, PDMS-BD-R moulding, and PDMS-without pattern (unpattern PDMS), respectively. SEM was carried out at 20,000× magnification. The nanopattern patterns formed in the PC-BD-R molding (Fig. 5A) and the PDMS-BD-R molding (Fig. 5B) are visible at a minimum magnification of 5000× to 20,000×.
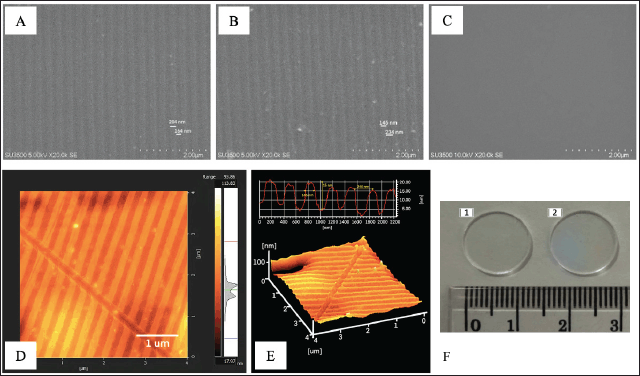 | Figure 5. Nanopattern substrate surface characteristics using SEM. Nano patterns on PC-BD-R using SEM after pre-processing at 20,000× magnification (4A). Nano patterns printed on PDMS-BD-R nanopattern at 20,000× magnification (4B). PDMS print without a pattern (control) at 20,000× magnification (4C). Inset scale = 2 and 5 µm. Characterization of PDMS-BD-R nanopattern using AFM, 2D imaging of PDMS-BD-R nanopattern using AFM (inset scale: 1 µm) (4D), and 3D imaging of PDMS-BD-R nanopattern using AFM (4E). (F) PDMS nanopattern morphology. (A) Grafting resulting from PDMS unpattern (flat) moulding. (F) Grafting resulting from PDMS-BD-R nanopattern molding. [Click here to view] |
The patterns formed on PC-BD-R and PDMS-BD-R nanopattern comprise nano topography in grooves and ridges. Dark areas depict the indentation depth, while lighter areas depict the top of the protrusion. On PC-BD-R, the groove width is approximately 164 ± 5.09 nm, and the protrusion width is around 204 ± 8.56 nm, while on PDMS-BD-R nanopattern, the groove width is approximately 234 ± 8.92 nm, and the width of the protrusion is around 145 ± 2.67 nm. SEM characterization results cannot predict the 3D shape of the nanopattern substrate along with the height or peak of the protrusions formed. Therefore, the PDMS-BD-R nanopattern was characterized using AFM to determine its three-dimensional structure in this research (Fig. 5D and E), AFM result shows the shape of the 3D PDMS-BD-R nanopattern with the peak height of the protrusion measured from the bottom of the indentation (depth), which is 15 ± 0.782 nm. Figure 5F above shows the physical shape of the PDMS substrate resulting from moulding without a pattern (Fig. 5F.1) and PDMS-BD-R nanopattern (Fig. 5F.2). The surface that has a nanopattern is characterized by the presence of rainbow-like color diffraction.
In vitro, the presence of biophysical signals derived from substrate topography has been extensively investigated as a means to regulate cell proliferation, control cell adhesion, guide cell motility, and direct MSC differentiation [32]. This approach is based on the fact that in vitro topographical features resemble the native ECM topography in vivo, including contours and patterns such as grooves, ridges, whorls, and pits, ranging from the nano- to microscale [33]. Consequently, cells can perceive biophysical signals from topographical features to regulate their behavior through FA interactions, triggering a series of cellular and molecular events known as mechanotransduction [34,35]. The nanopatterns fabricated in this study range in size from 100 to 300 nm, aligning with the findings of Rodríguez-Pereira et al. [36] reported that the RGD-Cys-D1 PLLA nanopatterned substrate (a substrate formed from a combination of dendrimer patterns and RGD, measuring 70 nm in diameter and 10 nm in height) was able to induce the formation of hAD-MSCs and hBM-MSCs aggregates/nodules, which serve as indicators of successful chondrogenesis. This process was accompanied by an increase in type II collagen synthesis [36].
WCA test
Plasma treatment on the PDMS substrate changes the physical character of the surface of the substrate, which was previously hydrophobic (Fig. 6A) to a substrate with a more hydrophilic surface (Fig. 6B). A decrease in the contact angle formed was seen on the PDMS substrate that was given plasma treatment, which previously had a ddH2O contact angle value of 85.426o (Fig. 6A) to 49.399o (Fig. 6B). Meanwhile, the PDMS substrate treated with plasma treatment and coated with spidroin extract provides high wettability with a ddH2O contact angle value of 14.562o (Fig. 6C). The role of both is to reduce the surface tension on the PDMS substrate so that water spreads more easily evenly.
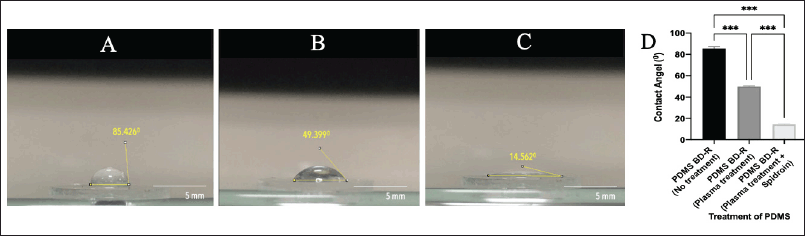 | Figure 6. A contact angle was formed on PDMS material coated with spider web extract. (A) PDMS-BD-R nanopattern material before plasma treatment, (B) after plasma treatment, and (C) after plasma treatment and coated with spidroin (spider web extract). (D) WCA analysis graph. The Bluray PDMS-BD-R nanopattern, treated with plasma treatment and spidroin extract coating, can reduce the contact angle 5–6 times. The smaller the contact angle formed; the more hydrophilic the material’s properties are. Data are mean ± SD with significance marker *** (p < 0.001). [Click here to view] |
Coating spider web extract on a PDMS substrate treated with plasma acts as an adsorbed protein that helps cells adhere well to the substrate. It is known that proteins adsorbed on substrate surfaces support cell attachment, cell spreading, and cytoskeletal organization much more than hydrophobic surfaces without adsorbed proteins [37]. A material is considered hydrophilic if its contact angle is less than 90° [38]. Furthermore, a smaller contact angle indicates enhanced cell attachment to the substrate, as it promotes better surface wettability [37].
Morphology of hWJ-MSCs cultured on PDMS-BD-R nanopattern
The morphology of the interaction between hWJ-MSCs cells and the nanopattern substrate was observed using SEM at 72 hours. Figure 7A shows the interaction between hWJ-MSCs cells on the PDMS-unpattern and PDMS-BD-R nanopattern surfaces. hWJ-MSCs cells cultured on PDMS-BD-R nanopattern coated with spidroin extract (Fig. 7A.3–4) provide a better picture of interactions with the substrate when compared with cells cultured on PDMS-unpattern (Fig. 7A.1–2). On the PDMS-BD-R nanopatterned substrate, cells exhibit enhanced growth and alignment along the defined pattern. The cell density is notably higher, and the cytoplasm elongates, revealing pronounced extensions of lamellipodia and filopodia. The cells are not round (passive) but are elongated as a sign that the cells are carrying out active movements. At this stage, the cell has experienced an advanced stage of initial cell contact with the substrate, known as cell spreading. Meanwhile, hWJ-MSCs cultured on unpatterned PDMS exhibited reduced cell density, with limited intercellular interactions and suboptimal contact with the substrate. Additionally, the cytoplasmic extensions of the cells were not fully developed, resulting in the absence of visible lamellipodia and filopodia.
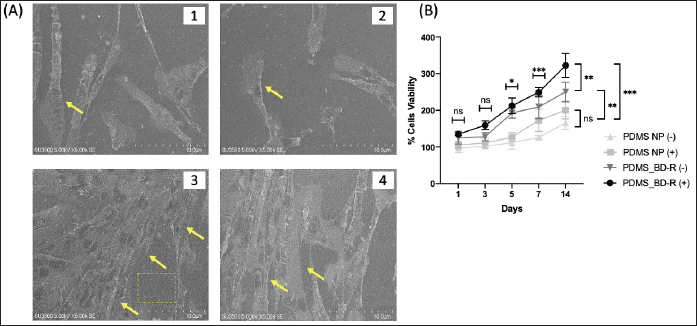 | Figure 7. Morphology of hWJ-MSCs on PDMS-BD-R nanopattern and unpatterned PDMS using SEM. (A.1-2) Morphology of hWJ-MSCs cells cultured on Unpattern PDMS coated with spidroin extract at 5,000× magnification. (A3-4) Morphology of hWJ-MSCs cells cultured on PDMS-BD-R Nanopattern coated with spidroin extract at 5,000× magnification. (B) Proliferation graph of hWJ-MSC cells cultured on PDMS-BD-R nanopattern and unpattern (NP) PDMS elastomers, whether coated with spidroin extract (+) or not (-). The proliferation of hWJ-MSC cells was measured using the MTT method on days 1, 3, 5, 7, and 14. Data are mean ± SD with significance markers * (p < 0.05) ** (p < 0.01) and * ** (p < 0.001). [Click here to view] |
The attachment of cells to nanopattern substrates is mediated by restructuring of the cytoskeleton as cells change shape, mostly through actin cross-linking, thereby changing the local fluidity of the cytoplasm [39]. Cells cultured on nanopattern surfaces have higher cytoskeleton tension than cells cultured on substrates with flat surfaces, thus affecting their interaction with the substrate [40]. Substrates with PDMS material have high hydrophobicity, making it difficult for cells to stick. This is where the adsorbed protein obtained from spidroin and plasma treatment facilitates the acceleration of cell attachment to the substrate. Trantidou et al. [37] described the role of protein adsorbed on the substrate as a cell accelerator to anchor and form FAs.
Proliferation of hWJ-MSCs cultured on PDMS-BD-R nanopattern
Figure 7B shows a graph of the proliferation of hWJ-MSCs cells cultured on two different substrate conditions, namely the PDMS-unpattern (PDMS-NP) and PDMS-BD-R nanopattern (PDMS-BD P), both coated with spider web extract (+) and not (-). There was no statistically significant difference in the levels of cell proliferation on days 1 and 3. An increase in hWJ-MSCs cell proliferation was only seen on days 5, 7, and 14, which was statistically significantly different. The appendix to the Supplementary Information section contains more thorough information on statistical differences. During the process of cell development, differentiation, and proliferation are coordinated activities. The success of the chondrogenic differentiation process is determined by the high cell proliferation exhibited by hWJ-MSCs. Similar to a study by Luo et al. [41], nanotubes topography has been shown to promote cell proliferation by modulating their adhesion behavior. According to Dexheimer et al. [42], during hMSC chondrogenesis, hWJ-MSCs cell proliferation and differentiation are closely synchronized. The rate and proliferation rate of expanded hWJ-MSCs showed a positive correlation to the development of chondrogenesis based on type II collagen accumulation, proteoglycan (GAG) deposition, DNA content, and the size of the chondrocyte cell nodules formed [43].
Matrix GAG abundance analysis
Figure 8A and B shows the quantitative and qualitative analysis of the abundance of the GAG proteoglycan matrix formed, respectively. hWJ-MSCs cells cultured on the PDMS-BD-R nanopattern had a different morphology than cells cultured on PDMS-unpattern whether coated with spidroin (+) or not (-). Statistically, GAG accumulation on day 7 between the treatment groups, whether coated with spidroin (+) or not (-), was not significantly different (Fig. 8A). GAG accumulation significantly differed between the treatment groups seen on day 14. However, the PDMS-BD-R nanopattern with and without spidroin coating was not significantly different but was significantly different from the treatment unpattern (PDMS NP). On the 21st day, the accumulation of GAG on the PDMS-BD-R nanopattern, whether coated with spidroin or not, significantly differed, with a p-value < 0.05. Meanwhile, compared to the treatment group without nanopatterns (PDMS NP), both with and without spidroin coating, they were significantly different, with a p-value < 0.001. This research is similar to earlier investigations wherein GAG matrix abundance increased on days 14 to 21 of the in vitro chondrogenesis process [12,16].
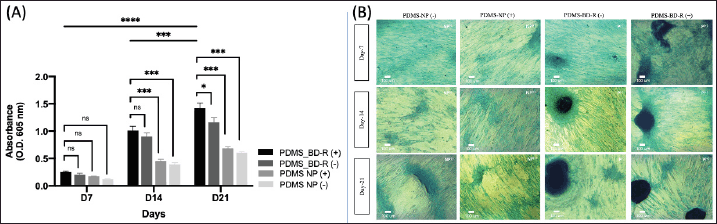 | Figure 8. (A) Quantification of matrix GAG abundance with Alcian Blue staining was measured over a period of 21 days at several points (days 7, 14, and 21). Data are mean ± SD with significance markers * (p < 0.05), ** (p < 0.01), *** (p < 0.001), and **** (p < 0.0001), total n = 4. (B) Morphology of hWJ-MSC cells with Alcian Blue staining cultured on PDMS elastomer substrate coated with spidroin extract (+) and without spidroin extract (-) on days 7, 14, and 21. NP = Unpattern PDMS. P = PDMS BD-R nanopattern. Inset scale = 100 µm, magnification 20×. [Click here to view] |
In Figure 8B, hWJ-MSCs cells cultured on the PDMS-BD-R nanopattern were able to form chondrocyte nodule aggregates starting from days 7 and 14. The color intensity and formation of chondrocyte nodules increased on days 14 and 21 in the PDMS-BD-R nanopattern treatment group. Meanwhile, the prospective cell aggregates that will form chondrocyte nodules in the PDMS-NP treatment were only formed on the 21st day but still needed to be completely formed. The nodules formed are a collection of hWJ-MSCs cells, which condense to form chondrocyte cell aggregates and secrete the ECM matrix. The nodules formed are also an indicator of the success of the chondrogenic differentiation process of hMSCs [42–44].
The condensed hMSCs are a prerequisite for successful chondrogenic differentiation. At this stage of cellular aggregation, hMSCs forming nodules (cell aggregates) undergo cellular communication facilitated by the expression of adhesion molecules such as N-cadherin, NCAM, or gap junctions [45]. Although this study does not molecularly confirm the presence of adhesion molecules, the formation of chondrocyte-like nodules, represents the successful completion of the chondrogenesis process [36,42]. The findings of this study align with those of Lagunas et al. [45] and Rodríguez-Pereira et al. [36] demonstrating that the RGD-Cys-D1 PLLA nanopatterned substrate (a substrate combining dendrimer patterns with RGD measuring 70 nm in diameter and 10 nm in height) successfully promotes the aggregation/nodule formation of hAD-MSCs and hBM-MSCs. This serves as a hallmark of successful chondrogenesis, accompanied by increased synthesis of type II collagen protein.
ICC analysis of PDMS-BD-R nanopattern
Based on the ICC results, Figure 9 below shows the type 2 collagen protein and SOX9 expression, respectively. The expression of type 2 collagen begins to be synthesized on day 7 and continues to increase until day 21, marked by a fluorescent green color. Differences were seen in the group of hWJ-MSCs cells cultured on the PDMS-BD-R nanopattern surface, which had a higher cell density. The cells began to experience perfect alignment following the pattern formed on day 7. After this treatment, the expression of SOX9 and type 2 collagen protein increased compared to the hWJ-MSCs cell group incubated on the PDMS-unpatterned surface.
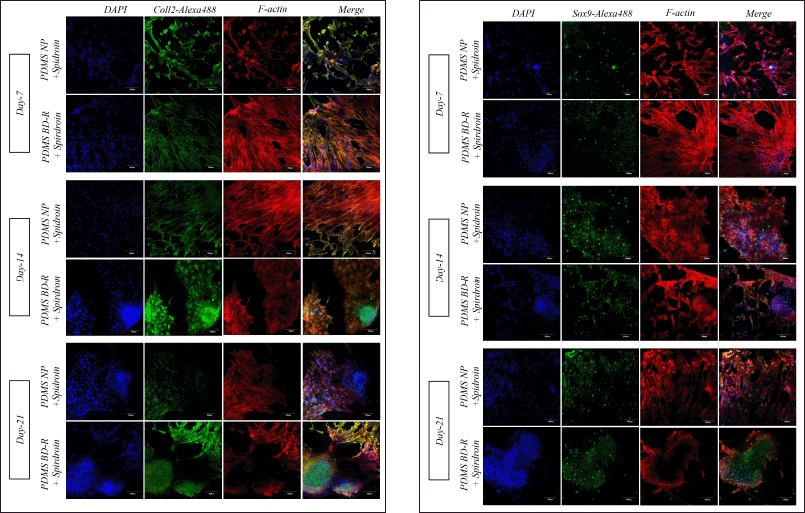 | Figure 9. Type II collagen and SOX9 expression with ICC on days 7, 14, and 21. Type II Collagen and SOX9 expression is marked in green, while the red color shows the actin protein and the blue color shows the cell nucleus. Scale: 200 µm. PDMS BD-R; PDMS nanopattern substrate. PDMS NP; PDMS substrate without a pattern (un-pattern). [Click here to view] |
On the 14th day, hWJ-MSCs cells cultured on the PDMS-BD-R nanopattern began to experience condensation to form chondrocyte nodules. Type 2 collagen synthesis is secreted more in the part of the cells that undergo condensation until it is visible, and the green color contrast is detectable until the 21st day. Meanwhile, SOX9 expression detected in the cell nucleus was visible starting on day 7 and increased along with the number of cells proliferating and differentiating until day 21. SOX9 expression is required as an initial step for hMSCs to condensate, proliferate, and differentiate into chondrocyte cells. According to Tew and Hardingham [46], SOX9 is an important transcription factor directly regulating type II collagen expression. Upregulation of SOX9 mRNA is considered with the prior expression of the COL2A1 gene and numerous other chondrogenic indicators [46]. MSCs become chondrocyte cells. During the development of the chondrogenesis process until the formation of hypertrophic chondrocytes, type 2 collagen protein, and SOX9 are the most abundant and dominant types of protein expressed so that they are widely used as biochemical markers for the continuation of the hMSCs chondrogenesis process [47,48].
To regulate cell function in vivo, biological and mechanical signals from surrounding cells and the ECM must be integrated [49]. In this research, the interaction between cells and the substrate surface is initiated with the role of protein coating from spider web extract (spidroin), which contains an RGD sequence that facilitates cell attachment to the substrate through integrin proteins [7]. The controlled geometrical features of nanopatterns, such as nano grooves and nano ridges on the PDMS-BD-R substrate, play a significant role in guiding stem cell behavior, influencing processes like adhesion, migration, proliferation, and differentiation.
The regulated geometrical cues of nanopatterns are related to the findings of a study that showed that minor changes in topography can offer necessary geometrical/physical signals in stem cell growth. The geometric cues that influence this research are patterns in the form of nano grooves and nano ridges that form on the surface. In several studies, it has been proven that geometrical cues regulated by topography engineering are able to influence the processes of adhesion, migration, proliferation, and differentiation of hMSCs [5,6]. Nanopatterns provide geometric signals and mechanical forces to cells, thereby influencing cell function and morphology. Both responses are transmitted as mechanical signals from the substrate to the nucleus, becoming a molecular process collectively known as mechanotransduction, the mechanical forces transmitted from the substrate to the nucleus impact cell function and morphology [9].
The synergistic interaction between specific nanopattern geometries (ridge/groove) and Spidroin protein can modulate cell differentiation through the mechanotransduction process. Mechanotransduction in this study involves the translation of mechanical signals into biochemical signals that affect the cytoskeleton and its interaction with the nucleus [49,50]. hWJ-MSCs cultured on PDMS-BD-R+Spidroin can sense the nanotopographic contours of the substrate through FA formation. This leads to an increased clustering of integrins and adhesion molecules around the cells, which determines the number and distribution of FA formed. The higher the FA formation, the more significant the changes in cytoskeletal organization and structure, particularly in inducing intracellular tension. The attachment of cells to the substrate activates a FA complex, which involves proteins like talin, vinculin, and focal adhesion kinase [51]. This leads to increased cytoskeleton tension, influencing cell differentiation. The formation of FA complexes due to the interaction of cells with nanopatterns causes tension in the cytoskeleton, which can ultimately influence cell differentiation.
The specific cellular signaling pathways activated in hWJ-MSCs upon interaction with the defined nanoscale ridge/groove patterns include integrin-mediated signaling and cytoskeletal reorganization. Based on Tsimbouri et al. [40], cells cultured on nanopattern surfaces have higher cytoskeleton tension than cells cultured on substrates with flat surfaces [52]. The high tension of the cytoskeleton is directly proportional to the number of FA complexes that will be formed so that, in the end, it can influence the shape of the cell nucleus. The more FA complexes that are formed, the more cytoskeleton contractility will increase through polymerization and crosslinking of actin filaments to induce stress fiber formation [40]. Stress fibers are a collection of actomyosin proteins consisting of actin filaments and myosin motor proteins as well as other components such as fimbrin, fascin, alpha-actinin, and filamin [53]. A research review by Barlian and Vanya [54] reported that cells cultured on nanopattern surfaces had high FA complexes so they were well able to form stress fibers. Sterss fiber generated intracellular tension through changes in cytoskeleton contractility will cause deformation of the cell’s nuclear membrane, thereby increasing nucleopore openings, which allows high levels of mRNA transport and protein translation [5].
On nanopatterned surfaces, the formation of FA complexes promotes higher cytoskeleton tension and the formation of stress fibers, which affect nuclear organization and epigenetic processes. These epigenetic changes activate genes related to chondrogenesis, such as COL2A1 and SOX9. The combination of nanopattern engineering and spidroin protein coating facilitates the chondrogenic differentiation of hWJ-MSCs without the need for biochemical regulators like growth factors or hormones. Based on the explanation above, the role of nanopatterns combined with spider web extract proteins is able to facilitate the induction process of chondrogenic differentiation of hWJ-MSCs cells without biochemical regulators such as growth factors/hormones. The nanopattern engineering in this research induces the mechanotransduction process, while spidroin helps attach hWJ-MSCs cells to the nanopattern through integrin-substrate interaction. The chondrogenic differentiation of hWJ-MSCs cells is most likely due to the interaction between hWJ-MSCs cells and nanopatterns, which induces the mechanotransduction process and ends in epigenetic modifications that activate several marker related to the chondrogenesis process.
CONCLUSION
PDMS-BD-R nanopatterns with ridge, groove, and depth patterns of 145 ± 2.67, 234 ± 8.92, and 15 ± 0.782 nm, respectively, successfully enhanced hWJ-MSCs chondrogenesis. The findings of this study were positively correlated with increases in chondrogenesis markers. Chondrogenic differentiation commences on days 14 to 21, as evidenced by an increase in the GAG matrix, the development of chondrocyte cell nodules, and the expression of collagen type II and SOX9 proteins. However, more studies are needed to investigate the molecular processes regulating the chondrogenic pathway caused by combining nanopatterns and spidroin protein coating.
ACKNOWLEDGMENTS
The authors would like to thank the Indonesia Endowment Fund for Education (Lembaga Pengelola Dana Pendidikan), Ministry of Finance of the Republic of Indonesia, for their support and for funding this research under grant number KET-5505/LPDP.4/2022. as well as PT Wadya Prima Mulia and Evident Singapore for providing access to the Confocal Laser Scanning Microscope, which was required for this study. The author is also grateful to Soraya Rahmanisa, M.Si and Rizka Musdalifah Amsar, M.Si to assist and guide the primary culture of hWJMSCs.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
Ethical approvals details are given in the ‘Materials and Method’ section.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Khalisha A, Puspitasari RL, Moegni KF, Rosadi I, Rosliana I. Profil Mesenchymal Stem Cell (MSC) Pasien Klinik Hayandra Pada Media Kultur Bersuplemen Menggunakan Flow Cytometry. Jurnal Al-Azhar Indonesia Seri Sains Dan Teknologi. 2018;4(4):195. CrossRef
2. de Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13(6):721–8. CrossRef
3. Somoza RA, Welter JF, Correa D, Caplan AI. Chondrogenic differentiation of mesenchymal stem cells: challenges and unfulfilled expectations. Tissue Eng Part B Rev. 2014;20(6):596–608. CrossRef
4. Ma Y, Ji Y, Huang G, Ling K, Zhang X, Xu F. Bioprinting 3D cell-laden hydrogel microarray for screening human periodontal ligament stem cell response to extracellular matrix. Biofabrication. 2015;7(4):044105. CrossRef
5. Cun X, Hosta-Rigau L. Topography: a biophysical approach to direct the fate of mesenchymal stem cells in tissue engineering applications. Nanomaterials. 2020;10:1–41. CrossRef
6. Wu C, Chin CSM, Huang Q, Chan HY, Yu X, Roy VAL, et al. Rapid nanomolding of nanotopography on flexible substrates to control muscle cell growth with enhanced maturation. Microsyst Nanoeng. 2021 Dec 1;7(1):89. CrossRef
7. Barlian A, Judawisastra H, Ridwan A, Wahyuni AR, Lingga ME. Chondrogenic differentiation of Wharton’s Jelly mesenchymal stem cells on silk spidroin-fibroin mix scaffold supplemented with L-ascorbic acid and platelet rich plasma. Sci Rep. 2020 Dec 1;10(1):19449. CrossRef
8. Kim J, Bae W, Kim YJ, Seonwoo H, Choung H, Jang K, et al. Directional matrix nanotopography with varied sizes for engineering wound healing. Adv Healthc Mater. 2017;6(19):1700297. CrossRef
9. Anene-Nzelu CG, Choudhury D, Li H, Fraiszudeen A, Peh KY, Toh YC, et al. Scalable cell alignment on optical media substrates. Biomaterials. 2013 Jul;34(21):5078–87. CrossRef
10. Wu C, Lin TG, Zhan Z, Li Y, Tung SCH, Tang WC, et al. Fabrication of all-transparent polymer-based and encapsulated nanofluidic devices using nano-indentation lithography. Microsyst Nanoeng. 2017;3(1):1–9. CrossRef
11. Tan J, Chan ZY, Lim PE, Koh JKH, Yong HS. A multigene approach to determine the molecular phylogeography of Argiope mangal and Argiope dang (Araneae: Araneidae) and their genetic relationships with the Argiope aetherea species group. Biochem Syst Ecol. 2016;69:115–23. CrossRef
12. Hernando A, Saputri DHA, Tan MI, Barlian A. Directing the chondrogenic differentiation of human Wharton’s jelly mesenchymal stem cells using spider silk-based micropattern. AIP Conference Proceedings, American Institute of Physics Inc.; 2021. CrossRef
13. Hofmann S, Knecht S, Langer R, Kaplan DL, Vunjak-Novakovic G, Merkle HP, et al. Cartilage-like tissue engineering using silk scaffolds and mesenchymal stem cells. Tissue Eng. 2006;12(10):2729–38. CrossRef
14. Wang Y, Kim UJ, Blasioli DJ, Kim HJ, Kaplan DL. In vitro cartilage tissue engineering with 3D porous aqueous-derived silk scaffolds and mesenchymal stem cells. Biomaterials. 2005;26(34):7082–94. CrossRef
15. Kluge JA, Rabotyagova O, Leisk GG, Kaplan DL. Spider silks and their applications. Trends Biotechnol. 2008;26(5):244–51. CrossRef
16. Barlian A, Saputri DHA, Hernando A, Khoirinaya C, Prajatelistia E, Tanoto H. Spidroin striped micropattern promotes chondrogenic differentiation of human Wharton’s jelly mesenchymal stem cells. Sci Rep. 2022 Dec 1;12(1):4837. CrossRef
17. Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. CrossRef
18. Secunda R, Vennila R, Mohanashankar AM, Rajasundari M, Jeswanth S, Surendran R. Isolation, expansion and characterisation of mesenchymal stem cells from human bone marrow, adipose tissue, umbilical cord blood and matrix: a comparative study. Cytotechnology. 2015 Oct 24;67(5):793–807. CrossRef
19. Pham LH, Vu NB, Van Pham P. The subpopulation of CD105 negative mesenchymal stem cells show strong immunomodulation capacity compared to CD105 positive mesenchymal stem cells. Biomed Res Ther. 2019 Apr 30;6(4):3131–40. CrossRef
20. Widowati W, Gunanegara RF, Rizal R, Widodo WS, Amalia A, Wibowo SHB, et al. Comparative analysis of Wharton’s Jelly mesenchymal stem cell (WJ-MSCs) isolated using explant and enzymatic methods. J Phys Conf Ser. 2019;1374:012024. CrossRef
21. Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini FC, Krause DS, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy. 2006 Aug;8(4):315–7. CrossRef
22. Samimi S, Maghsoudnia N, Eftekhari RB, Dorkoosh F. Lipid-based nanoparticles for drug delivery systems. In: Mohapatra SS, Ranjan S, Dasgupta N, Mishra RK, Thomas S, editors. Characterization and biology of nanomaterials for drug delivery: nanoscience and nanotechnology in drug delivery. Amsterdam, The Netherlands: Elsevier; 2018. pp. 47–76. CrossRef
23. Matusiak J, Grz?dka E. Stability of colloidal systems—a review of the stability measurements methods. Ann Univ Mariae Curie-Sklodowska Sect AA Chem. 2017 Dec 8;72(1):33. CrossRef
24. Honary S, Zahir F. Effect of zeta potential on the properties of nano-drug delivery systems-a review (Part 1). Trop J Pharm Res. 2013;12(2):255–64. CrossRef
25. Hack R. Viscosity. In: Bobrowsky PT, Marker B, editors. Encyclopedia of engineering geology. encyclopedia of earth sciences series. Cham, Switzerland: Springer; 2018. pp. 926–9. Available from: http://link.springer.com/10.1007/978-3-319-73568-9_308 CrossRef
26. Wen C, Yu C, Thirumalaivasan N, Hiramatsu H. 532-nm-excited hyper-Raman spectroscopy of amino acids. J Raman Spectrosc. 2021 Mar;52(3):641–54. CrossRef
27. Xiao L, Wang H, Schultz ZD. Selective detection of RGD-integrin binding in cancer cells using tip enhanced Raman scattering microscopy. Anal Chem. 2016 Jun 21;88(12):6547–53. CrossRef
28. Zhu G, Zhu X, Fan Q, Wan X. Raman spectra of amino acids and their aqueous solutions. Spectrochim Acta A Mol Biomol Spectrosc. 2011 Mar;78(3):1187–95. CrossRef
29. Pflüger F, Hernández B, Ghomi M. Vibrational analysis of amino acids and short peptides in hydrated media. VII. Energy landscapes, energetic and geometrical features of l -histidine with protonated and neutral side chains. J Phys Chem B. 2010 Jul 15;114(27):9072–83. CrossRef
30. Rygula A, Majzner K, Marzec KM, Kaczor A, Pilarczyk M, Baranska M. Raman spectroscopy of proteins: a review. J Raman Spectrosc. 2013;44:1061–76. CrossRef
31. Altomare L, Riehle M, Gadegaard N, Tanzi M, Farè S. Microcontact printing of fibronectin on a biodegradable polymeric surface for skeletal muscle cell orientation. Int J Artif Organs. 2010;33(8):535–43. CrossRef
32. McNamara LE, McMurray RJ, Biggs MJP, Kantawong F, Oreffo ROC, Dalby MJ. Nanotopographical control of stem cell differentiation. J Tissue Eng. 2010;1(1):120623. CrossRef
33. Keung AJ, Kumar S, Schaffer D V. Presentation counts: microenvironmental regulation of stem cells by biophysical and material cues. Annu Rev Cell Dev Biol. 2010;26:533–56. CrossRef
34. Cigognini D, Lomas A, Kumar P, Satyam A, English A, Azeem A, et al. Engineering in vitro microenvironments for cell based therapies and drug discovery. Drug Discov Today. 2013;18:1099–108. CrossRef
35. Jansen KA, Atherton P, Ballestrem C. Mechanotransduction at the cell-matrix interface. Semin Cell Dev Biol. 2017;71:75–83. CrossRef
36. Rodríguez-Pereira C, Lagunas A, Casanellas I, Vida Y, Pérez-Inestrosa E, Andrades JA, et al. RGD-dendrimer-poly(L-lactic) acid nanopatterned substrates for the early chondrogenesis of human mesenchymal stromal cells derived from osteoarthritic and healthy donors. Materials. 2020 May 1;13(10):2247. CrossRef
37. Trantidou T, Elani Y, Parsons E, Ces O. Hydrophilic surface modification of pdms for droplet microfluidics using a simple, quick, and robust method via pva deposition. Microsyst Nanoeng. 2017;3:16091. CrossRef
38. Jahangiri F, Hakala T, Jokinen V. Long-term hydrophilization of polydimethylsiloxane (PDMS) for capillary filling microfluidic chips. Microfluid Nanofluidics. 2020 Jan 1;24(1):2. CrossRef
39. Paddillaya N, Mishra A, Kondaiah P, Pullarkat P, Menon GI, Gundiah N. Biophysics of cell-substrate interactions under shear. Front Cell Dev Biol. 2019;7:251. CrossRef
40. Tsimbouri P, Gadegaard N, Burgess K, White K, Reynolds P, Herzyk P, et al. Nanotopographical effects on mesenchymal stem cell morphology and phenotype. J Cell Biochem. 2014 Feb;115(2):380–90. CrossRef
41. Luo J, Walker M, Xiao Y, Donnelly H, Dalby MJ, Salmeron-Sanchez M. The influence of nanotopography on cell behaviour through interactions with the extracellular matrix—a review. Bioact Mater. 2022;15:145–59. CrossRef
42. Dexheimer V, Frank S, Richter W. Proliferation as a requirement for in vitro chondrogenesis of human mesenchymal stem cells. Stem Cells Dev. 2012 Aug 10;21(12):2160–9. CrossRef
43. Zhu L, Skoultchi AI. Coordinating cell proliferation and differentiation. Curr Opin Genet Dev. 2001;11(1):91–7. CrossRef
44. Nazempour A, Quisenberry CR, Abu-Lail NI, Van Wie BJ. Enhancing adipose stem cell chondrogenesis: a study on the roles of dexamethasone, transforming growth factor β3 and ascorbate supplements and their combination. J Stem Cell Therapy Transplant. 2017;1:28–51. CrossRef
45. Lagunas A, Tsintzou I, Vida Y, Collado D, Pérez-Inestrosa E, Rodríguez Pereira C, et al. Tailoring RGD local surface density at the nanoscale toward adult stem cell chondrogenic commitment. Nano Res. 2017 Jun 1;10(6):1959–71. CrossRef
46. Tew SR, Hardingham TE. Regulation of SOX9 mRNA in human articular chondrocytes involving p38 MAPK activation and mRNA stabilization. J Biol Chem. 2006;281(51):39471–9. CrossRef
47. Lefèvre T, Auger M. Spider silk as a blueprint for greener materials: a review. Int Mater Rev. 2016;61(2):127–53. CrossRef
48. Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dynam. 1997;209(4):377–86. CrossRef
49. Martino F, Perestrelo AR, Vinarský V, Pagliari S, Forte G. Cellular mechanotransduction: from tension to function. Front Physiol. 2018;9:824. CrossRef
50. Ross TD, Coon BG, Yun S, Baeyens N, Tanaka K, Ouyang M, et al. Integrins in mechanotransduction. Curr Opin Cell Biol. 2013;25(5):613–8. CrossRef
51. Biggs MJP, Dalby MJ. Focal adhesions in osteoneogenesis. Proc Inst Mech Eng H. 2010;224(12):1441–53. CrossRef
52. Tsimbouri P. Adult stem cell responses to nanostimuli. J Funct Biomater. 2015 Jul 16;6(3):598–622. CrossRef
53. Miller CJ, Harris D, Weaver R, Ermentrout GB, Davidson LA. Emergent mechanics of actomyosin drive punctuated contractions and shape network morphology in the cell cortex. PLoS Comput Biol. 2018;14(9):e1006344. CrossRef
54. Barlian A, Vanya K. Nanotopography in directing osteogenic differentiation of mesenchymal stem cells: potency and future perspective. Future Sci OA. 2022;8(1):FSO765. CrossRef
SUPPLEMENTARY MATERIAL
The supplementary material can be accessed at the link here: [https://japsonline.com/admin/php/uploadss/4566_pdf.pdf]