INTRODUCTION
Natural antimicrobial agents (AMAs) can be acquired by plants (herbs) or their byproducts, animal origin, bacterial origin, algae, and mushrooms. However, several research findings indicate that the consumption or use of crude or derivatives of herbals or plants or their byproducts is superlatively significant compared to other various sources in modern treatment for various diseases including antimicrobial activity [1].
Natural products (NPs) have been used for over 80 years and continue to save countless lives annually. Most of the antibiotics on the market today are derived from NP classes that were identified over half a century ago [2,3]. A major hazard to public health and a global issue is the emergence of antimicrobial resistance (AMR) and estimated 700,000 deaths annually and expected to reach 20 million by 2050 [4]. Historically, natural substances have been essential in the discovery and creation of antimicrobial drugs and alternatives to synthetic medications and the antibacterial effectiveness varies depending on their structural variation [5]. The rapid evolution of resistant bacterial strains has compelled a re-evaluation of these systems despite a decline in interest in recent years [6]. The various benefits of plant-derived NPs over modern allopathic medications are presented in Figure 1. Lists of antibacterials are given in Tables 1–4.
According to Nathan and Cars (2014), the 1930s through 1960s were the “golden era” of antibiotic development. Antibiotics were regarded as the “wonder drug” in the middle of the 20th century [7]. In India, almost 20,000 recognized medicinal plants were found [8–10]. Figure 2 pie chart displays the section on the use of herbal remedies and their components in various medical systems for various therapeutic goals. It is known that microbes that cannot be grown in a typical laboratory setting account for 95%–99% of the entire microbiome [11–14].
THE RISK-BENEFIT RATIO OF HERBAL THERAPY
India, which contains more than 17,000 species that have been identified and 7,500 plants having therapeutic use, is the ideal place to find numerous traditional compounds. Allopathic medications can cure a variety of illnesses, including cancer. Nevertheless, due to their exorbitant cost and unanticipated, potential AMR and fatal side effects, many people are turning back to natural sources like herbal medicines, which have negligible or nonexistent side effects [14]. Numerous benefits or advantages of herbals, or their formulations, are based on diverse evaluations of the literature [15–33], and are indicated in Figure 1. Particularly in relation to AMR, the NPs have long been a valuable source of antibiotics since they have special chemical structures and methods of action that can be favorable over synthetic antibiotics. Their advantage in pharmacokinetics, toxicity, and bioavailability is multifarious. For example, a naturally occurring substance obtained from Bacillus polymyxa, colistin, is potent against Gram-negative bacteria resistant to many drugs. Still, poor gastrointestinal absorption calls for parenteral treatment. This emphasizes that although some natural antibiotics have strong efficacy, their bioavailability may be restricted, so different administration techniques are necessary.
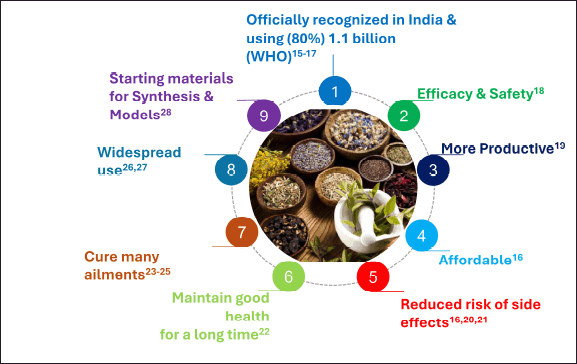 | Figure 1. The various benefits of plant derived natural products over modern allopathic medications. [Click here to view] |
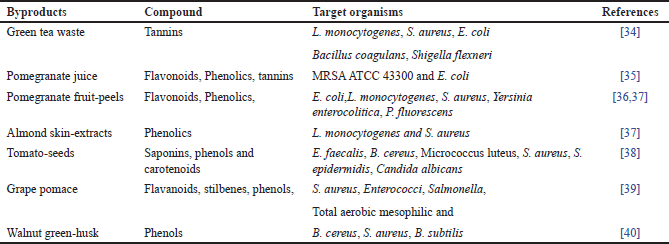 | Table 1. Antimicrobials from plant-by-products. [Click here to view] |
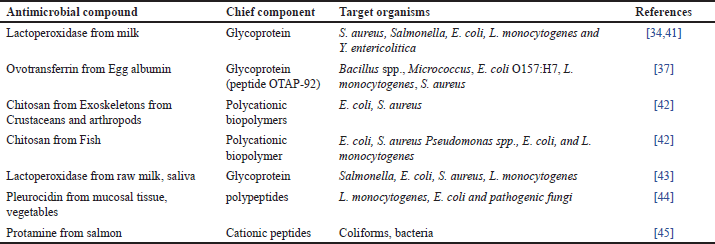 | Table 2. Antimicrobials from animal origin. [Click here to view] |
 | Table 3. Antimicrobials from bacterial origin. [Click here to view] |
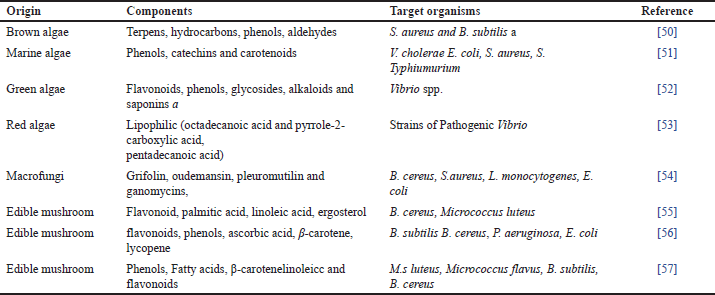 | Table 4. Antimicrobials from algae and mushrooms. [Click here to view] |
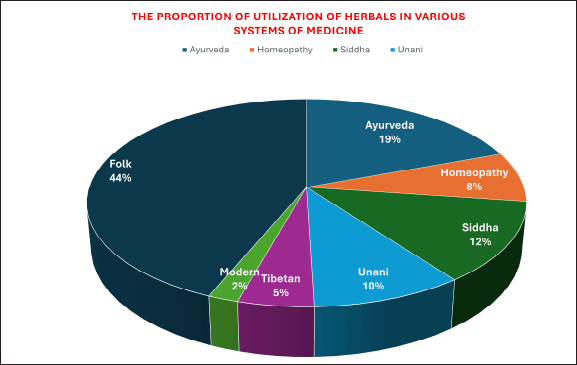 | Figure 2. Use of herbal remedies and their components in various medical systems. [Click here to view] |
Different natural antibiotics have different toxicity profiles. Because of their nephrotoxicity and neurotoxicity, polymyxins—including colistin—are only used to treat serious infections when no other treatment works. On the other hand, some NPs are less hazardous, which makes them appropriate for a wider range of uses. Natural chemicals’ varied structures make it possible to choose and optimize molecules with fewer negative effects. Natural compounds have unique mechanisms that can bypass established resistance pathways. For example, fosmidomycin inhibits 1-deoxy-D-xylulose 5-phosphate reductoisomerase, a critical enzyme in the non-mevalonate route of isoprenoid biosynthesis that is lacking in humans but found in some bacteria and parasites. This selectivity lowers the risk of cross-resistance with human metabolic pathways. Furthermore, natural compounds can improve the efficacy of existing antibiotics by blocking bacterial efflux pumps or virulence factors, restoring the action of antibiotics that bacteria have developed resistance to.
ANTIMICROBIAL PRODUCTS FROM ENVIRONMENTAL SOURCES
AMA can be acquired from plants (herbs) or their byproducts, animal origin, bacterial origin, algae, and mushrooms. Some of these AMA are listed in Tables 1–4 and the basic cultivation approach for novel antimicrobials (Fig. 3) and modern developments in antimicrobial discovery from NPs are shown in Figures 4 and 5. A great deal of research has been done recently to identify natural antimicrobials that can halt the progression of bacteria and fungi in food, thus increasing its quality and prolongation of shelf-life [58]. Consequently, there has been a hunt for antimicrobials that come from different natural sources. Numerous studies on plants for AMAs have shown the usefulness of substances in food applications, as well as the variables affecting this efficacy [59]. Thus, the presence of hydrophobic components in essential oils (EOs), such as phenolic compounds, may be the cause of the antibacterial action of plant extracts.
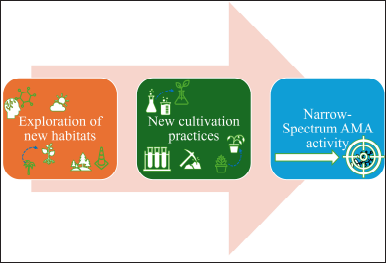 | Figure 3. The basic cultivation approach for novel antimicrobials. [Click here to view] |
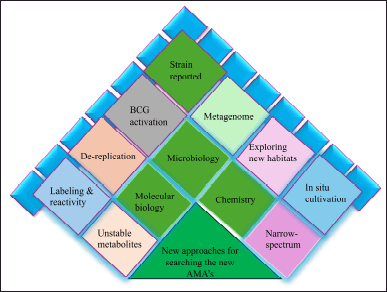 | Figure 4. Modem approaches in the selection of new AMAs from natural products. [Click here to view] |
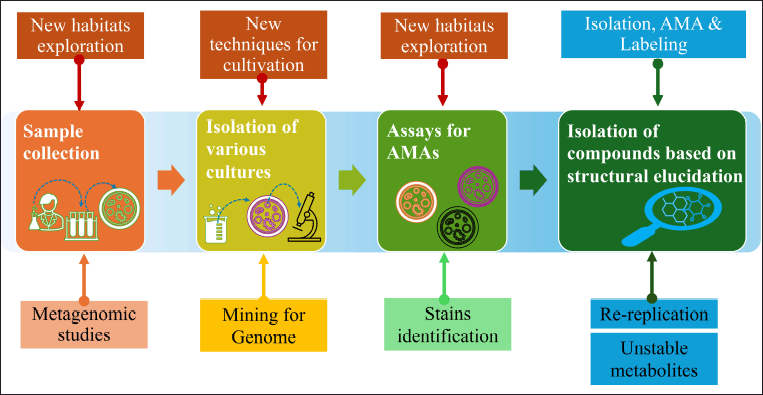 | Figure 5. Schematic representation of modern developments in antibiotic discovery. [Click here to view] |
Antimicrobials from plants
The antibacterial activity of these substances varies according to changes in their structure and chemical makeup [60]. Since ancient times, plant-derived chemicals found in herbs and spices have been utilized as preservatives, traditional medicines, and food flavorings. The major studies on plants as AMA were published [60]. In this part, we provide an overview of these compounds’ antibacterial activity, largely based on their structural characteristics. Secondary metabolites have antibacterial qualities that protect against spoilage and harmful microorganisms, among other advantages [60].
Plants have major classes of chemicals called phenolics/acids, quinones, saponins, flavonoids, tannins, terpenoids, alkaloids, and others that exhibit antimicrobial activity [61]. Among the most varied classes of secondary metabolites, phenolic compounds exhibit significant structural diversity. Since the (–OH) groups of phenols can interact with bacterial cell membranes to break membrane assemblies and cause seep out of the cellular components, and it is believed that these groups have an inhibitory effect. The –OH group found in phenolic compounds is crucial to the antibacterial activity of thymol and carvacrol [62]. Active groups like –OH encourage electron delocalization, which in turn serves as a proton exchanger and lessens the incline across the bacterial cell’s cytosolic membrane. This will lead to the depletion of the ATP pool, the proton motive force collapsing, and ultimately cell death. The very last enzyme in the oxidative phosphorylation pathway to trigger ATP synthesis is called ATP synthase (also known as F-ATPase or F0F1-ATP) the enzyme responsible for synthesizing ATP from its precursor, ADP and inorganic phosphate (Pi) using electrochemical energy in the oxidative phosphorylation pathway. The collapse of the proton motive force and the depletion of ATP reduce the activity of this enzyme, contributing to the antibacterial effects. Additionally, with an ever-rising need for novel antibiotics due to the phenomenon of antibiotic resistance, serious research into creative workarounds for this problem is highly encouraged, and as this review made clear, one of the promising therapies is the ATP synthase inhibitors [63].
The efficacy of EOs can be enhanced by combining molecules with varying biochemical capabilities, as well as by their lipophilic qualities, the potency of functional groups, or water solubility, in addition to their chemical makeup. This process involves the breakdown of membranes by lipophilic chemicals, which inhibit phosphorylation, and translocation of protein electron transport [64,65]. Certain antimicrobial plant compounds are listed in Table 1.
Numerous secondary metabolites found in plants, including flavonoids, alkaloids, terpenoids, and tannins, have been shown to possess antibacterial qualities in vitro. The collection of microorganisms that coexist in and around a plant and work together to create a microbial ecosystem is known as the plant microbiome. All types of microbiomes have intimate interactions with both living and non-living elements of their surroundings. The antimicrobial chemicals found in medicinal plants may offer a substantial clinical benefit in the treatment of resistant microbial strains and may work through different processes than currently used antimicrobials to suppress the growth of bacteria, fungus, viruses, and protozoa. Emetine, quinine, and berberine are still very efficient tools in the battle against microbial infections. Historically, plants have been a good source of anti-infective medicines. For instance, a variety of Gram-positive and Gram-negative bacteria can be inhibited by the crude extracts of curry, ginger, sage, mustard, garlic, cinnamon, and basil [60–62].
Antimicrobials from plants byproducts
A significant number of by-products, such as pomace fruits, peels, seeds, pulps, leftover flesh, and husks, are frequently produced throughout the food preparation process. Even though these by-products are usually regarded as waste, some studies have shown that the husks, peels, seeds, and kernels are potentially valuable sources of phenolic compounds (polyphenols, tannins, and flavones) as well as numerous other bioactive substances with a variety of functions, including antimicrobial activity [66,67] and certain antimicrobial plant compounds are listed in Table 1. Pomace is a valuable by-product that is made up of seeds and stems. It is high in phenolic compounds, minerals, polyphenols, flavonoids and non-flavonoids, and dietary fiber [68]. It has been demonstrated that 10% ethanolic extract of grape pomace inhibits the growth of Salmonella, Staphylococcus aureus, and Enterobacteriaceae [39].
Fruit peels’ antibacterial properties are widely known. For instance, pomegranate fruit peels are frequently utilized in herbal medicines to treat a variety of illnesses, and it has been demonstrated that extracts from pomegranate fruit peels can stop the growth of several pathogens, like Yersinia enterocolitica, Bacillus cereus, S. aureus, and Listeria monocytogenes [69]. However, when it came to Gram-negative bacteria, the extract worked better at a concentration (0.1%) against Pseudomonas species and less well at the same concentration against Salmonella Typhimurium and Escherichia coli [70]. Antimicrobial action is displayed by flavonoids, tannins, phenolics, and active inhibitors found in peels [71]. It has been shown that the peel of bergamot, a byproduct of the EOs, is effective against both Gram+ve Bacillus subtilis and Gram-negative pathogens like E. coli and S. enterica. For the bergamot ethanolic fraction, the MIC ranged from 0.2 to 0.8 mg/ml [72].
Quinine antibacterial pursuit against S. aureus, yeast (C. albicans), E. coli, and P. aeruginosa [73]. Mango kernel extracts have demonstrated an inclusive range of antibacterial activity against several foodborne pathogens. Mango seed kernel at 3,000 ppm showed antibacterial efficacy against E. coli [74]. This antibacterial activity is explained by the presence of polyphenolics [75].
Studies on tomato seed extracts have demonstrated antibiotic efficacy against a variety of bacteria (G- and G+) and fungi (B. cereus, C. albicans, E. faecalis, S. aureus, and Staphylococcus epidermidis) [38]. Like this, husks of fruits as byproducts that can be utilized as a convenient way to obtain naturally occurring bioactive chemicals with antibacterial qualities [40]. It was discovered that almond skin fractions had antibacterial activity in the range (0.25–0.5mg/ml) hostile to S. aureus and L. monocytogenes. Legume hulls have the potential to be a unique natural addition because of their antibacterial activity against pathogenic bacteria and common food deterioration [76].
The antibacterial properties of coffee pulp were examined about rancidity and microbiological development. Very few investigations into the antibacterial properties of coffee byproducts, such as coffee pulp and tea trash [77]. These investigations have yielded valuable insights into the application of various fruits and vegetable by-products (peels, seeds, pulps, and husks) as naturally occurring antimicrobials in food. Furthermore, the residue generated by the industrial sector may be employed as edible antimicrobial films or integrated into antimicrobial packaging [38].
Tropical plant food by-products’ antibacterial properties are reviewed. Banana peel and coconut root extracts have strong antimicrobial properties. By-product alkaloids, phenolics, and saponins exhibit potent antibacterial properties. Numerous studies look at the microbiological properties of a few secondary metabolites, such as tannins, terpenoids, alkaloids, and phenolics, that are typically found in plant-food by-products. However, the list of compounds to look for in by-products is nearly endless because all of these metabolites have a large number of subclasses of active chemicals. As previously mentioned, pure chemicals that are isolated from plants are often characterized to accurately determine their antibacterial activities. Research on the antibacterial qualities of by-products has characterized the profiles of active chemicals and investigated the antimicrobial activity of various extracts. The efficient utilization of by-products in animal production farms is made possible by an accurate understanding of their active-compound composition. However, using by-products is a significant problem. Only a small number of publications have explained how these substances work by removing their active ingredients [38,77].
Antimicrobials of animal origin
Specific potential antimicrobials of animal origin are listed in Table 2. It has been discovered that the iron-binding glycoprotein lactoferrin (Lf) in milk has antibacterial pursuit against a variety of bacterial and viral infections [78]. Lf has been used as an anti-bacterial activity in the US [79,46]. It has been claimed that Lf possesses antibacterial endeavors against food-borne pathogens such as Carnobacterium, E. coli, L. monocytogenes, and Klebsiella by taking up the microbial iron [80].
Chitosan is one of several naturally occurring antimicrobials that have invited a lot of attention for use in food products. Crustaceans and arthropods naturally contain the polycationic biopolymer chitosan in their exoskeletons [80] and chitosan demonstrated to be a viable commercial replacement for acid-soluble against S. aureus, L. monocytogenes, B. cereus, E. coli, S. Typhimurium, and Shigella dysenteries [81,82]. The enzyme lysozyme is found naturally in both animal milk and avian eggs. It is commonly agreed as safe (GRAS) to add directly to food. The bacteriolytic enzyme known as white lysozyme from hen eggs is frequently utilized as a protective for meat and meat products, milk, fish, fruits, and vegetables [42]. It has also been well-documented for its usage as an antibacterial in food items [83].
The antibacterial activity of lysozyme stems from its capacity to hydrolyze the beta-1, 4-links between N-acetylglucosamine and N-acetylmuramic acid in the peptide-glycan of the cell wall [46] with an inhibition (-) of 19.75nm and 17.37mm, respectively; Lysozyme demonstrated the strongest antibacterial activity against Saccharomyces cerevisiae and Listeria innocua [84]. It has been shown that bioactive substances originating from milk, such as whey protein and casein have a variety of uses, including being antibacterial [85]. αS1-and αS2-casein have been found to contain antibacterial peptides [86] against B. subtilis, Diplococcus pneumoniae, Staphylococcus species, Sarcina species, and Streptococcus pyogenes [87].
Ciprofloxacin and enrofloxacin are among the residual antimicrobials found in the organs of birds. Amoxicillin in tilapia and chloramphenicol in rui are examples of residual antibiotics in fish. AMR can be brought on by antimicrobial residues in meat, milk, eggs, and fish. Animal parts that are most likely to come into touch with environmental pathogens are often where AMP are located. As a result, they are present in the bone marrow and testes, on the skin, ear, and eye, and on epithelial surfaces such as the tongue, trachea, lungs, and gut. The transmission of antibiotic-resistant bacteria to humans, immunopathological effects, allergies, mutagenicity, nephropathy (gentamicin), hepatotoxicity, reproductive abnormalities, bone marrow toxicity (chloramphenicol), and even carcinogenicity are just a few of the adverse effects that these residues may have. The immune system relies heavily on antimicrobial proteins. By immediately eliminating infections or impeding their ability to replicate, they help the body fight them. Interferons, the complement system, iron-binding proteins, and antimicrobial proteins are the four primary categories of antimicrobial compounds [42–87].
Antimicrobials of bacterial origin
Specific bacterial origin AMA are listed in Tables 3 and 4. Numerous chemicals with antibacterial action are fabricated by microbes, particularly lactic acid bacteria (LAB). Among these, it has been demonstrated that the proteinaceous substances known as bacteriocins, like nisin, impede the expansion and maturation of other microbial species. Comparably, another popular broad-spectrum antibacterial agent is reuterin made from glycerol in some strains of L. reuteri. Reuterin also works well against a variety of spoilage and harmful microbes.
More than 50 countries throughout the world utilize nisin, the only bacteriocin that has been licensed for use as an antibacterial. Produced by Lactoccocus lactis, it exhibits efficacy against food-associated Gram+ve bacterium [88]. Nisin acts as a bactericide against Gram+ve foodborne pathogens and spoilage bacteria, such as M. luteus, S. aureus, and B. cereus [89]. It accomplishes this by penetrating the cytoplasmic membrane, which results in the loss of membrane potential and the release of intracellular metabolites [49].
Reuterin (β-hydroxypropionaldehyde) is a chemical that demonstrates antibacterial action against a wide range of spoilage and foodborne-pathogens. Reuterin is an excellent choice for a food biopreservative due to its high-water solubility, resistance to heat, proteolytic and lipolytic enzymes, and stability across a broad pH range [90]. Reuterin and nisin together have been shown to have a synergistic effect on L. monocytogenes and S. aureus. Gram+ve (S. aureus, L. monocytogenes) and Gram-ve (E. coli, S. Typhimurium) bacteria were significantly reduced in number by reuterin from L. reuteri DPC16 [91].
Algae and mushrooms have been reported to be natural sources of bioactive compounds with a wide range of biological activity in recent years. These compounds include antimicrobial, antioxidant, cytotoxic, antifouling, anti-inflammatory, and antimitotic activities. On the other hand, not significant research has been done has been done to evaluate the antibacterial activity of mushrooms and algae. Therefore, the wide variety of algae and mushroom species may provide a possible trace for new antimicrobials [92].
Reportedly shown compounds include phenols, terpenoids, phlorotannins, acrylic acid, steroids, ketones and alkanes, cyclic polysulphides, and fatty acids with bactericidal properties [93]. The mushrooms are frequently used as food and have been found to have antibacterial qualities [94]. Recently reported on the potential use of various genus of macro-fungi as a potential food supplement and a good resource of natural antibiotics compounds. Extracts from Australian basidiomycetous macrofungi were evaluated against both Gram-ve (P. aeruginosa) and Gram+ve (B. cereus, L. monocytogenes) bacteria. At 10mg/ml, the ethanol extracts of macro fungus that were examined stopped one or more of these infections from growing [54]. The existence of certain secondary metabolites like volatile compounds, gallic acids (GAs), phenols, free fatty-acids (FFAs), and their spinoffs of mushrooms also be sensible for the antibacterial action [95].
The use of nanotechnology in the preparation of natural compounds has garnered the attention of numerous researchers in recent years [96]. This technology has the potential to enhance antimicrobial stability and prevent the growth of foodborne pathogens in various food systems by being applied directly, as coating, or as packaging [97]. Many studies have reported on the use of nanotechnology to deliver antimicrobials. However, because food components are so complex, there has been very little research done on nanoparticles as antimicrobials (AMAs). The potential application of liposomal nanoparticles to augment nisin’s antibacterial activity against L. S. aureus and monocytogenes was demonstrated and investigated the antibacterial activity of solid lipid nanoparticles loaded with nisin and free nisin [97,98].
Antimicrobials come from bacteria. Antimicrobial substances (such as bacteriocin, organic acids, diacetyl, and hydrogen peroxide) that are effective against pathogenic bacteria are frequently produced by LAB. The most often utilized antibacterial agents are isopropyl alcohol, n-propanol, and ethyl alcohol. Although it works as a disinfectant as well, methanol is rarely utilized due to its severe toxicity. Alcohols have the ability to impede the growth of certain bacteria, including Salmonella, Staphylococcus aureus, and Escherichia coli. In short, typically cationic peptides having an amphiphilic structure, AMPs readily bind and interact with the cellular membranes of bacteria, fungi, viruses, and other pathogens. Both Gram-positive and Gram-negative bacteria can manufacture bacterial AMPs, also known as bacteriocins, through ribosome synthesis in order to eradicate competing organisms. In order to combat the growing antibiotic resistance of different diseases, bacterial AMPs are essential and may even be a substitute for inefficient antibiotics. Bacteriocins are extremely specialized antibacterial substances that target certain bacterial infections due to their limited spectrum of action. The two primary classes of bacteriocins that are produced by Gram-positive and Gram-negative bacteria are their classification, their mechanisms of action, the beneficial effects they can have on the human body, their drawbacks, and their potential as an antibiotic substitute [92–98].
ANTIMICROBIALS AND MECHANISM OF RESISTANCE
Like the phenomena of tolerance observed in higher organisms, antimicrobial-resistance is the inability of a micro-organism to respond to an antibiotic [99] which persistently presents a noteworthy public health concern concerning mortality and financial loss. Action plans have been devised by health authorities in several nations, including India, to contain it. It is crucial to recognize the role that it plays. By 2025, AMR alone is predicted to hamper the 10 million population than cancer and car accidents 700,00 deaths annually [100]
International bodies, the World Health Organization (WHO), and various other stakeholders are visualizing the global rise in AMR which has been declared an urgent priority, and numerous European world leaders have outlined their action plans for containing AMR. Real attempts are being made to defeat this AMR [101]. National Action Plan (NAP) of India for AMR has also been developed to understand that AMR is a complex issue that necessitates a multifaceted approach to contain [102,103].
MECHANISM OF AMR
A schematic presentation of the mechanism of development of AMR is presented in Figure 6 and several pathways for prevention of AMR are also shown in Figure 7. Certain AMAs have always caused resistance in some microorganisms as they do not have the target location or metabolic pathway that the specific medication affects. There are various sources for attaining AMR such as Natural resistance. For instance, gram-negative bacilli unaffected by Penicillin-G, anerobic is not inhibited aminoglycosides, aerobic organisms not affected by metronidazole and M. Tuberculosis is insensitive to tetracyclines. Acquired resistance is developed by a pathogenic organism (which was sensitive earlier) due to the consumption of AMA over a period. This is a serious clinical issue that can occur with any bacterium. However, both the antibiotic and the microbe have a role in the development of resistance. Some bacteria, like staphylococci, coliforms, and tubercle bacilli, are infamous for developing resistance quickly (sulfonamides resistance by Gonococci).
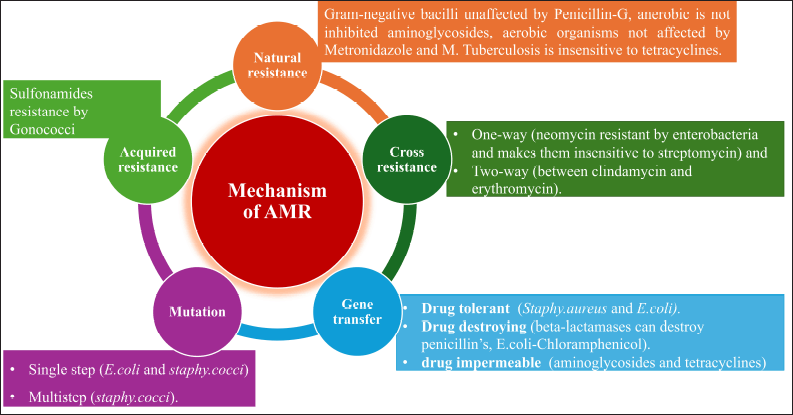 | Figure 6. Mechanism development of antimicrobial resistance. [Click here to view] |
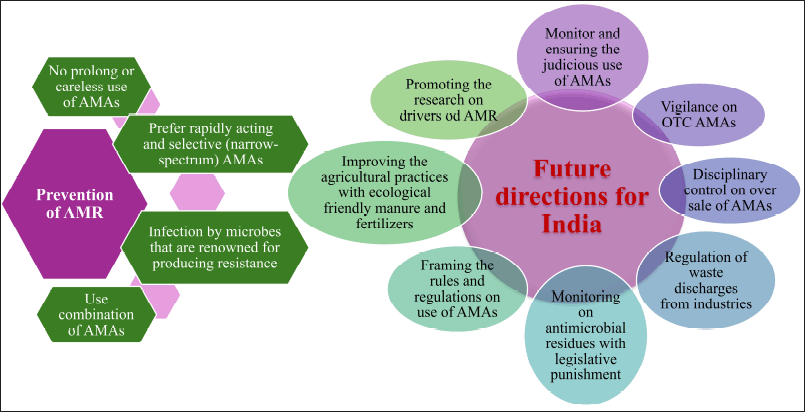 | Figure 7. Possible ways for prevention of AMR and Future directions for India. [Click here to view] |
The AMR may be developed by mutation or gene transfer. Mutation is a spontaneous, random genetic alteration that is both stable and heritable in microorganisms. A few mutant cells exist in every sensitive population of a microorganism, and these cells need larger concentrations of AMA to be inhibited. These are kept apart and allowed to grow as the AMA destroys the sensitive cells. Consequently, when a single antitubercular medication is administered, for example, it may eventually appear that a sensitive strain has been replaced by a resistant one. This phenomenon, known as vertical transfer of resistance, is often of lower grade and occurs rather slowly. A mutation may be developed by a single step (E. coli and S. cocci) or multistep (S. cocci).
Gene transfer is called infectious resistance in which a resistance gene transferred to another organism is called horizontal transfer of resistance. This might be caused by various mechanisms such as conjugation, transduction, and transformation. However, resistant organisms can be classified as: a) drug tolerant (loss of affinity to the AMA, e.g., Staphy.aureus and E. coli). b) Drug destroying (beta-lactamases can destroy penicillin’s, E. coli-Chloramphenicol). c) Drug impermeable (several hydrophilic antibiotics require certain transport mechanisms or enter the bacterial cell through channels made by proteins known as “porins.” The resistant strains might lose these, e.g., aminoglycosides and tetracycline) and d) Cross-resistance (cross-resistance is the development of resistance to one AMA that transfers resistance to another AMA that the organism has not been exposed to). This is more frequently observed in medications that are related chemically or mechanistically. This may be one-way (neomycin resistant by enterobacteria and makes them insensitive to streptomycin) and two-way (between clindamycin and erythromycin) [99,104].
PROBLEM STATEMENT FOR INDIA’S REPORTED AMR RATE
AMR containment relies on the interconnection of humans, animals, and environmental characteristics [105]. Another concern is the paucity of investigated data, which makes it challenging to determine the exact incidence and spread of AMR in India and hinders national comparisons. Out of the 2152 AMR research publications published by Indian institutes, 48.3% concentrated on people, while only 3.3% on animals, 4.2% on the environment, and 0.5% addressed One Health. The remaining research was focused on novel agents, miscellaneous, editorials, and diagnostics [106].
THE CURRENT SCOPE OF THE AMR CHALLENGES IN INDIA
As stated in the “AMR Scenario Report for India, 2017” under the auspices of the Indian government, more than 70% of isolates of Klebsiella pneumoniae, E. coli, and A. baumannii, as well as over half of all isolates of Pseu. aeruginosa, were tolerant to fluoroquinolones and 3rd gen. cephalosporins. While only about 35% of E. coli and P. aeruginosa exhibited resistance to the piperacillin-tazobactam medication combination, 65% of K [105].
The increasing prevalence of carbapenem-resistant A. baumannii led to the frequent usage of colistin as a last-resort antibiotic which reached 71%. In India, there has also been a rise in colistin-resistance except for the 4.1% [106]. Genes and microorganisms resistant to antibiotics have been identified in some Indian water sources. When residential waste and hospital effluent were the management plant’s input, domestic water alone, and hospital effluent alone, respectively, the rate of separation of E. coli tolerant to 3rd-gen cephalosporins was 25, 70, and 95% [107].
AMR CHALLENGES IN INDIA
India is called “the world’s AMR capital” [108]. Disparate newly emerging multi-drug-resistant (MDR) pathogens, which pose significant research and therapeutic challenges. India is still working to fight off long-standing foes including drug-resistant cholera pathogens, malaria, and tuberculosis. Conditions like hunger, overcrowding, poverty, and illiteracy exacerbate the problem [109]. The general populace is often prevented from obtaining medical advice due to their restricted access to healthcare and lack of understanding about infectious diseases [110].
Due to this, antimicrobial drugs are frequently self-prescribed without a doctor’s expertise on dosage and length of therapy. Because of this, people frequently do self-medication without discussing with a doctor about the correct dose and length of treatment leads to MDR [111]. The amount of trash produced by these businesses has increased in tandem with the growth of the pharmaceutical industry. This trash enters water bodies and acts as a constant source of AMR in the environment due to lax oversight and a lack of enforcement [110,112]. The utilization of antimicrobial chemicals such as pesticides and insecticides in the agricultural sector is another significant challenge, notwithstanding the paucity of available data on the subject [113].
INDIA’S ENVIRONMENTAL AMR DRIVERS
In the risk assessment for AMR, Chereau et al. [114] have demonstrated that although environmental sources of AMR may contribute minimally to developed nations, they pose a reasonable to elevated hazard in evolving nations of Southeast Asia, including India, because of multiple co-factors related to the overall event. The schematic presentation of AMR drivers and preventive measurements is shown in Figure 8.
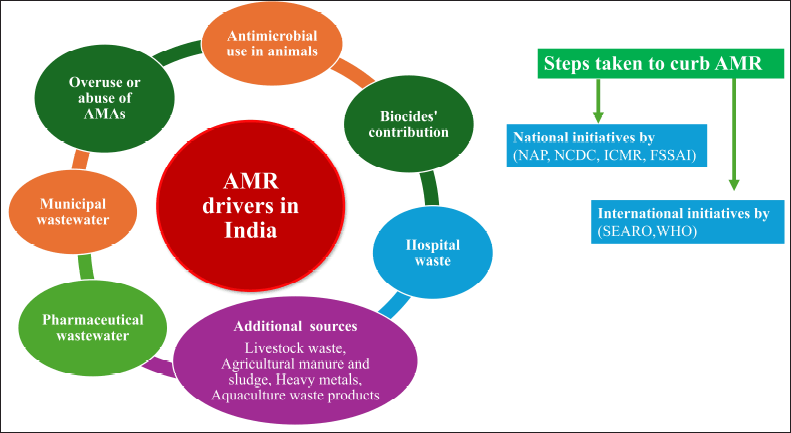 | Figure 8. AMR drivers in India and steps taken to curb AMR. [Click here to view] |
The major AMR environmental drivers include the following:
Overuse or abuse of antimicrobial substances
Abuse or prolonged use of antibiotics in India contributes to AMR in a greater extent. In 2010 alone, each person consumed 10.7 antibiotic units [115]. The amount of antibiotics sold at retail increased by 23% between 2000 and 2010 [116]. Roughly half of the antibiotics that are taken by mouth are eliminated by the body intact through urine and feces. Defecating in the open, as has been done for decades in India [117], results in the soil and after absorbing antibiotics or their residues. Nearly 35% of the population is exposed to drinking water contaminated with feces [118] which means that this environmental factor has a major role in the advancement of AMR.
AMR promoted using antimicrobial in animals
India is a significant manufacturer of animal goods for the world market, including meat, meat products, and farmed fish. By 2030 [119], there is predicted to be a 312 percent increase in this sector. Antimicrobial drugs are frequently utilized to boost productivity and prevent illnesses in these farmed animals [120]. India is the world’s fourth-largest user of antibiotics for use in animals, behind the US, China, and Brazil [116].
Antibiotic residues have been reported from food animal products in India, including milk and chicken flesh [108,121]. Future research examining the same is desperately needed, as there is no data available to characterize the domestic picture. The International Food Safety and Quality Network Standards have set strict restrictions regarding the use of antibiotics in food animals (IFSQNS) [110,122].
Biocides’ contribution to AMR
The fact that biocides and AMA share resistance pathways, which facilitates their co-selection, is another crucial factor. The co-location of both resistance genes on the plasmid49 results in an eight-fold increased tolerance to oxacillin for S. aureus when it is resistant to the biocide benzalkonium chloride. The biocide market experienced a 40% expansion on a global scale between 1992 and 2007 [110]. While data on biocide usage in India is scarce, it is believed to be substantial [123].
Pharmaceutical wastewater as a source of AMR
Ciprofloxacin levels were reported to be 28 and 31 mg/l in the wastewater of one of the Indian pharmaceutical firms [124].
Municipal wastewater
Water bodies in the vicinity are exposed to this potentially “AMR-rich” municipal wastewater (20%–30%). The Mutha River in Pune, India, has genes that are resistant to even the most powerful antibiotics. These genes are found in the sediments close to the city, and they are thought to have originated from home and municipal sewage waste. The concentration of these genes is thirty times higher than normal [125].
9.6 Hospital waste
Antimicrobial waste gets generated in substantial amounts by healthcare facilities and other healthcare facilities, either directly from residual or unused pharmaceuticals or indirectly from patient discharges. It has been reported to what extent hospital pollutants in India contained traces of tinidazole, sulfonamide, and fluoroquinolone [126]. Given that healthcare facilities are the locations with the highest levels of antibiotic practice, it is expected that these waterways will have the highest concentration of tolerant bacteria and their genetic material. Antimicrobial concentrations from hospital wastewater plants in India were high enough to affect bacterial strains and cause genotoxic alterations [127]. 80%–85% of antibiotic residues can be successfully removed from hospital wastewater by processing it appropriately before final disposal, according to research [128].
Additional AMR sources
Livestock waste [129], agricultural manure and sludge [130], heavy metals [123], and aquaculture waste products [131].
ACTIONS TAKEN TO REDUCE THE AMR
International efforts
To plan and adopt strategies to address AMR in the region, the Southeast Asian Regional Office (SEARO) held multiple conferences. One such historic gathering took place in 2011 in India, Jaipur, where the health ministers of all the participating nations—including India—vowed to embrace the Jaipur Declaration and implement stringent AMR policies using the WHO’s 2014 Report [100]. India and the WHO worked collaboratively following the publication of this report. A Global Action Plan was launched after the 68th World Health Assembly resolved to incorporate the idea of One Health into the battle against AMR. All Member countries, including India, agreed to frame their NAPs for AMR by 2017. Notwithstanding the fact that the WHO identified AMR as the Flagship Attention area for SEARO [100], the Indian Medical Association in 2015 initiated an awareness drive [132] with the aim of educating physicians and raising public awareness [100].
Nationwide initiatives
An NAP for the nation was prepared by the Core Working Group on AMR, which was established by the Indian government. The NAP has identified six strategic priorities, all of which have considered the possibility of AMR occurring in the environment either straight or implicitly [133,134]. A plan for 5 years also outlines how each strategic priority’s assigned interventions, activities, and results are expected to be fulfilled. A list of the acceptable amounts of antimicrobial waste in foods made from fish, poultry, and animals was published by the Food Safety and Standards Authority of India (FSSAI) in June 2017 [135].
The Indian Council of Medical Research and the National Center for Disease Control, respectively, introduced the AMR monitoring platforms of Indian health agencies in 2013 and 2014 to determine the approximate scope of AMR [101]. According to the National Health Policy 2017, “pharmacovigilance including prescription audits inclusive of antibiotic usage - in the hospital and community” and “a rapid standardization of guidelines regarding antibiotic use, limiting the use of antibiotics as OTC medications, banning or restricting the use of antibiotics as growth promoters in animal livestock” are among the demands made [134]. In India, creative solutions and mass media are required to address such sociocultural concerns. We suggest the following possible ways for the prevention of AMR and future directions for India is shown in Figure 7.
CURRENT RESEARCH AND FUTURE PERSPECTIVES OF NATURAL ANTIMICROBIALS
In current scenarios of research and development, screening for antimicrobials with broad-spectrum activity often leads to toxic and re-isolation issues, and the “Waksman platform” of traditional phenotypic screening is depicted in Figure 3. Hence, screening needs prolonged cultivations (for isolating the axenic cultures) and re-sources-consumption activity guidance isolation of antimicrobials which cannot be possible by fast high-throughput screening techniques. Even though random investigation occasionally provides some interesting results a discovery of a new class of lipoglycopeptide antibiotic Gausemycin A/B against gram+ bacteria. Gausemycins A and B are cyclic peptides with a unique peptide core and several remarkable structural features, including unusual positions of D-amino acids, lack of the Ca2+ binding Asp-X-Asp-Gly (DXDG) motif, tyrosine glycosylation with arabinose, presence of 2-amino-4-hydroxy-4-phenylbutyric acid (Ahpb) and chlorinated kynurenine (ClKyn), N-acylation of the ornithine side chain. These major components of the peptide antibiotic family have pronounced activity against Grampositive bacteria [136]. The structure is shown in Figure 9. Nacubactam is an experimental drug that is part of the diazabicyclooctane (DBO) class and is intended to treat infections brought on by Enterobacteriaceae that are resistant to carbapenem (CRE). It works in two ways: it directly targets penicillin-binding protein 2 (PBP2) in Enterobacteriaceae and inhibits serine β-lactamases (classes A, C, and some D enzymes). This dual action has direct antibacterial effects in addition to shielding companion β-lactam drugs from degradation. Clinical trials are being conducted on nacubactam in conjunction with antibiotics such as meropenem, cefepime, and aztreonam to treat severe bacterial infections brought on by MDR organisms, including complicated UTIs.
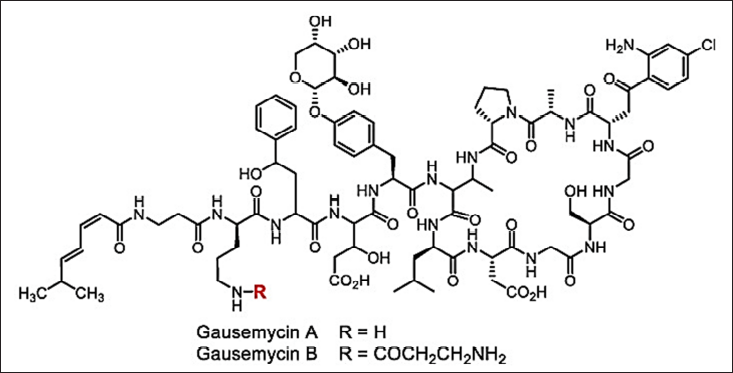 | Figure 9. Structure of Gausemycins. [Click here to view] |
In this research, we emphasize the key developments that get around the restrictions of phenotype screening both methods and outcomes of recent investigations. The new research approach evolves into multidisciplinary research, schematically with three main divisions such as microbiology, chemistry, and molecular biology as shown in Figure 4.
Isolating the natural antimicrobials by traditional methods will help in selecting only a small number of existing diversities [137]. The new cultivation approach for novel antimicrobials is shown in Figures 1–3. Exploring novel habitats, and new cultivation techniques (co-cultivation [138–141]. In-situ cultivation [142–147], microtechnology [148–150], and a new approach for narrow-spectrum antimicrobials [151,152], compounds with more selective effect are drawing increasing attention since they enable access to a portion of this mysterious microbiome, changing the paradigm [153]. The iChip technology with diffusion chambers of stainless steel and 0.03µm pore size to isolate the new antibiotic non-ribosomal peptide “teixobactin” from beta-proteobacterium-Eleftheria terrae that binds to lipid II and lipid III, essential precursors in cell wall biosynthesis. Teixobactin has notably prevented the development of observable resistance in infections, which represents a major breakthrough in the discovery of antibiotics. Teixobactin-analogue in complex with Lipid II in lipid membranes. The structure provides a wealth of information on the pharmacophore and demonstrates that teixobactins balance specific and fuzzy binding to recognize a maximal number of targets and minimize the likelihood of resistance development. Furthermore, our structure explains the critical sequence of D- and L-amino acids that enables a clear-cut separation of hydrophilic and hydrophobic sidechains and thereby also provides a molecular rationale why certain positions do not tolerate substitutions of hydrophobic residues by hydrophilic residues and vice versa [143] (Fig. 10). The potential of phage therapy in combating AMR was highlighted when researchers discovered a bacteriophage from Merri Creek, Melbourne, that was able to eradicate the superbug Klebsiella pneumoniae.
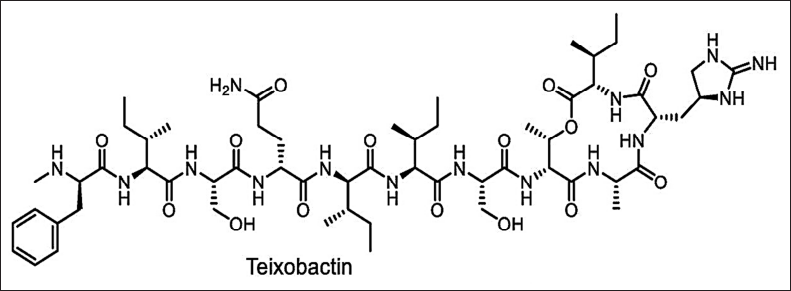 | Figure 10. Structure of Teixobactin. [Click here to view] |
Microtechnology (microarrays, microencapsulation, micromechanical, and microfluids) can also be applied to develop new antimicrobials by reducing time and resources [149,150]. The broad-spectrum antimicrobials have several disadvantages such as stability and bacterial resistance. Hence, new approaches to finding new narrow-spectrum activity antibiotics will avoid these limitations. In the year 2016, the key concept of finding the narrow-spectrum activity was projected by Brown and Wright [153] which is inspired by the example of Fidaxomicin and Hygromycin-A (Fig. 11). These new approaches direct the study of stain-specific and bacteriocins antimicrobials as next-generation products with narrow-spectrum activity (Fig. 12). Thus, the narrow-spectrum antimicrobials reduce the AMR and side effects [154].
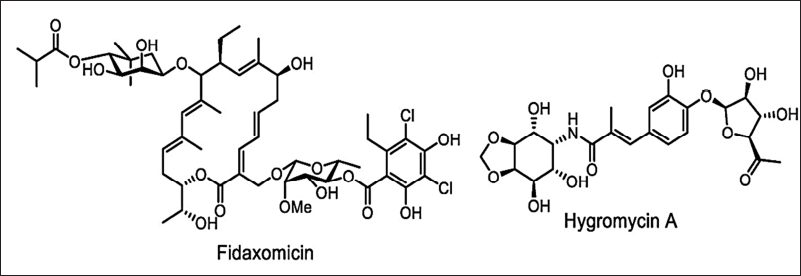 | Figure 11. Structure of Fidaxomicin and Hygromycin-A. [Click here to view] |
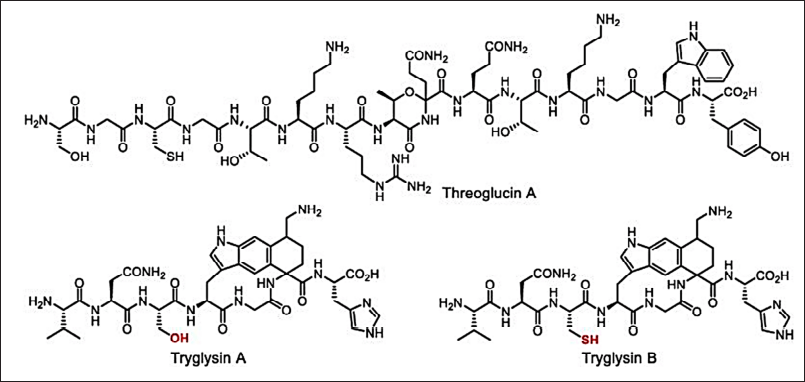 | Figure 12. Narrow-spectrum antimicrobials: threoglucin A and tryglusins A and B. [Click here to view] |
New genomic engineering (genome mining/metagenesis) and molecular biological approaches help in developing new antimicrobials [36–161]. The modern development in antimicrobial discovery is depicted in Figures 4 and 5. The clinically approved novel lipopeptide antibiotics, daptomycin discovered by a metagenomic study (Fig. 13). The metabolomics-guided identification represents a promising frontier in antimicrobial discovery. By leveraging comprehensive metabolite profiling and utilizing extensive databases like METLIN and ECMDB, researchers can uncover novel bioactive compounds. Addressing challenges related to data complexity and standardization will further enhance the efficacy of this approach in combating AMR.
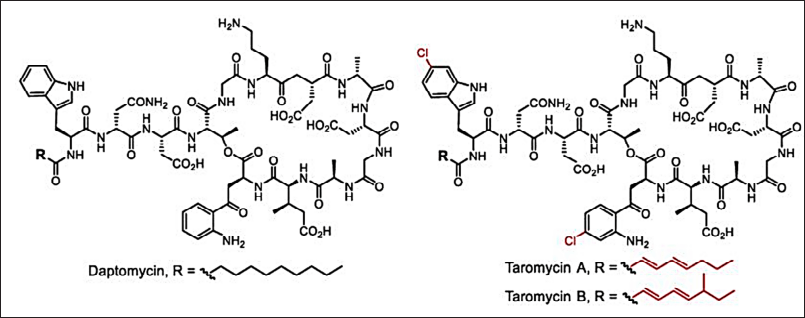 | Figure 13. Clinically approved novel lipopeptide antibiotics, daptomycin and taromycins [Click here to view] |
Recent research has revealed that proteins in Sydney rock oyster blood can kill bacterial infections and improve the efficacy of some medicines, so derived AMA is for antibacterial medicines derived from marine life, this finding creates fresh opportunities. Researchers are going back over past remedies like Lemnian Earth (LE), an old Greek medical clay. Modern research indicates that LE’s mix of particular clays and fungus could help gut bacteria, thereby providing a possible basis for modern gut health treatments. Molecular “De-Extinction”: Synthetic biology and artificial intelligence have made it possible to rebuild antibacterial compounds from extinct organisms. Scientists have created compounds from woolly mammoths and Neanderthals, for example, which have demonstrated encouraging antibacterial effects and offer fresh models for the development of drugs. The production of nanostructured surfaces influenced by natural antibacterial mechanisms has been made easier by the use of nanostructured bactericidal surfaces in bioengineering. Black silicon surfaces have bactericidal activity, as evidenced by research headed by Elena P. Ivanova, which mechanically ruptures bacterial cells upon contact. These biomimetic nanostructures provide a new class of mechanoresponsive antibacterial materials by mimicking the physical antibacterial characteristics found in insect wings. These surfaces could be used in implants and medical equipment to lower the risk of infections linked to biofilms.
The new “transcription-translation in One” (TTP) was described recently [162] and reporter-guided mutation selection was created for the novel metabolites [163,164] is shown in (Fig. 14). However, the establishment of receptor stains and mechanism-guided isolation is the advanced research for the elucidation of antimicrobials based on the mechanism of action (MoA) [165–167] The schematic representation of this screening strategy is shown in Figure 15.
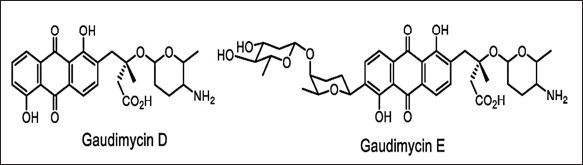 | Figure 14. Structures of gaudimycins D, E discovered by means of RGMS. [Click here to view] |
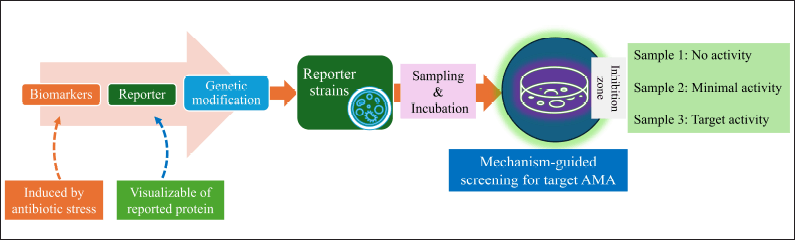 | Figure 15. Reporter strain-assisted screening strategy. [Click here to view] |
AI-Powered Screening for Antimicrobial Research
Predictive Modeling: By examining chemical structures and biological data, machine learning algorithms can forecast the antibacterial activity of natural substances, expediting the drug discovery process.
Virtual Screening: AI makes it possible to virtually screen enormous collections of NPs, effectively locating substances that may be effective against particular infections.
Finding novel Antibiotics: By using artificial intelligence (AI) to sift genomic data, researchers have found novel antibiotics from uncharted microbial sources.
LIMITATIONS AND CHALLENGES
Resistance development
Though new drugs like teixobactin show promise, there is still reason for worry about resistance development. Reducing this risk depends on constant observation and sensible use.
Regulatory and ethical considerations
Treatments like bacteriophage therapy are subject to ethical and regulatory issues, especially when it comes to their application in Western medicine. To overcome these obstacles, thorough clinical studies and standardized procedures must be established.
Translational barriers
Although ancient medicines and proteins derived from marine sources exhibit promise in vitro, obstacles including toxicity, bioavailability, and large-scale production must be addressed before these discoveries can be turned into clinically useful treatments.
Data quality and integration
The availability of thorough, high-quality datasets is a prerequisite for the use of AI in drug discovery. Strong data curation and validation are essential since incomplete or biased data can result in inaccurate predictions.
CONCLUSION
The development of antibiotic-resistant microorganisms emphasises how vital it is to create novel antimicrobials. However, the development of new drugs cannot proceed successfully or efficiently until appropriate drug leads are identified. In this assessment, we listed the main tactics for addressing the current problems with the development of natural antibiotics. The question, “How do we make antibiotics great again?” cannot be satisfactorily answered by any of these approaches. Additionally, the basic procedure remains unchanged and is essentially similar to the traditional phenotypic screening (often referred to as the “Waksman platform”). Every technique displayed enhances one or more phases of the traditional discovery paradigm. Using these developments could lead to a large increase in the rate of new antibiotic discovery. Combining several datasets (such as genomics and metabolomics) has been shown to produce new and valuable information. Further data integration, however, necessitates advanced instrumentation, processing power, and technology. It may become feasible to use natural compounds made from plants or their byproducts, algae, and mushrooms as novel antimicrobials. Utilizing these items may potentially be a more cost-effective way to make antimicrobials. Therefore, more research is required to identify the most effective antibacterial dosages that can be used without needlessly changing any sensory qualities. Therefore, developing a more practical and effective delivery strategy for these naturally occurring antibacterial compounds remains essential. Before being used in the healthcare industry, natural AMAs must also be evaluated for safety and potential health hazards in order to maximize the benefits for the patient.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Baranova AA, Alferova VA, Korshun VA, Tyurin AP. Modern trends in natural antibiotic discovery. Life. 2023;13(5):1073.
2. Bernal FA, Hammann P, Kloss F. Natural products in antibiotic development: is the success story over?. Curr Opin Biotechnol. 2022;78:102783.
3. Atanasov AG, Zotchev SB, Dirsch VM. Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov. 2021;20:200–16.
4. WHO. Antimicrobial resistance. Geneva: WHO; 2023.
5. Moloney MG. Natural products as a source for novel antibiotics. Trends Pharmacol. Sci. 2016;37(8):689–701.
6. Gyawali R, Ibrahim SA. Natural products as antimicrobial agents. Food Control. 2014;46:412–29.
7. Nathan C, Cars O. Antibiotic resistance--problems, progress, and prospects. N Engl J Med. 2014;371(19):1761–3.
8. Pandey MM, Rastogi S, Rawat AKS. Indian herbal drug for general healthcare: an overview. Internet J Altern Med. 2008;6:1–3.
9. Narayana A, Subhose V. Standardization of Ayurv?edic formulations: a scientific review. Bull Indian Inst Hist Med Hyderabad. 2005;35(1):21–32.
10. Belay B, Belachew B, Habitamu D. Review on application and management of medicinal plants for the livelihood of the local community. J Resour Dev Manage. 2016;22:33–9.
11. Salam N, Xian WD, Asem MD, Xiao M, Li WJ. From ecophysiology to cultivation methodology: filling the knowledge gap between uncultured and cultured microbes. Mar Life Sci Technol. 2021;3:132–47.
12. Mu DS, Ouyang Y, Chen GJ, Du ZJ. Strategies for culturing active/dormant marine microbes. Mar Life Sci Technol. 2021;3:121–31.
13. Moore BS, Carter GT, Brönstrup M. Editorial: are natural products the solution to antimicrobial resistance? Nat Prod Rep. 2017;34(7):685–6.
14. Laxminarayan R, Duse A, Wattal C, Zaidi AK, Wertheim HF, Sumpradit N, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13(12):1057–98.
15. WHO. WHO Traditional medicine strategy: 2014–2023. Essential medicines and health products 2013. Geneva: WHO; 2013.
16. Tilburt JC, Kaptchuk TJ. Herbal medicine research and global health: an ethical analysis. Bull World Health Organ. 2008;86(8):577–656.
17. Martins E. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front Pharmacol. 2013;4:177.
18. Yuan R, Lin Y. Traditional Chinese medicine: an approach to scientific proof 30 and clinical validation. Pharmacol Ther. 2000;86:191–8.
19. Gagnier JJ, Boon H, Rochon P, Moher D, Barnes J. Reporting randomized controlled trials of herbal interventions: an elaborated CONSORT statement. Ann Intern Med. 2006;144:364–7.
20. Lam TP. Strengths and weaknesses of traditional Chinese medicine and Western medicine in the eyes of some Hong Kong Chinese. J Epidemiol Community Health 2001;55:762–5.
21. Mi MK, Soobin J, Jeeyoun J. Herbal medicines for metabolic diseases with blood stasis. Medicine 2019;98:8:e14543.
22. Haidan Y, Qianqian M, Li Y, Guangchun P. Traditional medicine and modern medicine from natural products. Molecules. 2016;21:559.
23. Joshi K, Ghodke K, Patwardhan B. Traditional medicine to modern pharmacogenomics: Ayurveda Prakriti type and CYP2C19 gene polymorphism associated with the metabolic variability. Evid Based Complement Alternat Med. 2011;2011:249528.
24. Akinyemi O, Oyewole SO, Jimoh KA. Medicinal plants and sustainable human health: a review. Horticult Int J. 2018;2(4):194?195.
25. Debas TH, Laxminarayan R, Straus SE. Complementary and alternative medicine. In: Jamison DT, Breman JG, Measham AR, et al. Disease control priorities in development countries. New York, NY: Oxford University Press, pp. 1281–91, 2nd ed; 2006.
26. Kala CP. Assessment of species rarity. Curr Sci. 2004;86(8):1058–9.
27. Iris FFB, Wacht S. Herbal medicine: biomolecular and clinical aspects. 2nd ed. Milton Park: Taylor and Francis; 2011.
28. Cooper EL. CAM, eCAM, bioprospecting: the 21st century pyramid. Evid Based Complement Alternat Med. 2005;2(2):125–7.
29. Gavaghan H. Koop may set up new centre for alternative medicine. Nature. 1994;370(6491):591.
30. Fatemeh JK, Zahra L, Hossein AK. Medicinal plants: past history and future perspective. J Herbmed Pharmacol. 2018;7(1):1–7.
31. Jon CT, Kaptchuk TJ. Herbal medicine research and global health: an ethical analysis. Bull World Health Organ. 2008;86:594–9.
32. WHO. WHO Global report on traditional and complementary medicine. Geneva: WHO; 2019.
33. Chandrakant K, Arun G, Satyajyoti K, Shefali K. Drug discovery from plant sources: an integrated approach. Ayu. 2012:33(1):10–9.
34. Sung SH, Kim KH, Jeon BT, Cheong SH, Park JH, Kim DH, et al. Antibacterial and antioxidant activities of tannins extracted from 1539 agricultural by-products. J Med Plants Res. 2012;6:3072–9.
35. Reddy MK, Gupta SK, Jacob MR, Khan SI, Ferreira D. Antioxidant, antimalarial and antimicrobial activities of tannin-rich fractions, ellagitannins and phenolic acids from Punica granatum L. Planta Med. 2007;73:461–7.
36. Bauman KD, Butler KS, Moore BS, Chekan JR. Genome mining methods to discover bioactive natural products. Nat Prod Rep. 2021;38:2100–29.
37. Agourram A, Ghirardello D, Rantsiou K, Zeppa G, Belviso S, Romane A, et al. Phenolic content, antioxidant potential and antimicrobial activities of fruit and vegetable by-product extracts. Int J Food Prop. 2013;16:968–79.
38. Taveira M, Silva LS, Vale-Silva LS, Pinto EN, Valentão PC, Ferreres F, et al. Lycopersicon esculentum seeds: 1547 an industrial byproduct as an antimicrobial agent. J Agric Food Chem. 2010;58(17):9529–36.
39. Sagdic O, Ozturk I, Yilmaz MT, Yetim H. Effect of grape pomace extracts obtained from different grape varieties on microbial quality of beef patty. J Food Sci. 2011;76(7):M515–M521.
40. Oliveira I, Sousa A, Ferreira IC, Bento A, Estevinho L, Pereira JA. Total phenols, antioxidant potential and antimicrobial activity of walnut (Juglans regia L.) green husks. Food Chem Toxicol. 2008;46(7):2326–31.
41. Perez KL, Taylor TM, Taormina PJ. Competitive research and development on antimicrobials and food preservatives. Microbiol Res Dev Food Ind. 2012; 2012:109.
42. Tiwari BK, Valdramidis VP, O’Donnell CP, Muthukumarappan K, Bourke P, Cullen P. Application of natural antimicrobials for food preservation. J Agric Food Chem. 2009;57(14):5987–6000.
43. Ko K, Mendonca A, Ahn D. Influence of zinc, sodium bicarbonate, and citric acid on the antibacterial activity of ovotransferrin against Escherichia coli O157: H7 and Listeria monocytogenes in model systems and ham. Poult Sci. 2008;87:2660–70.
44. Burrowes O, Hadjicharalambous C, Diamond G, Lee TC. Evaluation of antimicrobial spectrum and cytotoxic activity of pleurocidin for food applications. J Food Sci. 2004;69:646–52.
45. Potter R, Truelstrup Hansen L, Gill TA. A. Inhibition of foodborne 1417 bacteria by native and modified protamine: Importance of electrostatic interactions. Int J Food Microbiol. 2005;103:23–34.
46. Juneja VK, Dwivedi HP, Yan X. Novel natural food antimicrobials. Annu Rev Food Sci Technol. 2012;3:381–403.
47. Siamansouri M, Mozaffari S, Alikhani FE. Bacteriocins and lactic acid bacteria. J Biol. 2013;2:227–34.
48. Gong H, Meng X, Wang H. Plantaricin MG active against Gram-negative bacteria produced by Lactobacillus plantarum KLDS1. 0391 isolated from “Jiaoke”, a traditional fermented cream from China. Food Control. 2010;21:89–96.
49. Lucera A, Costa C, Conte A, Del Nobile MA. Food applications 1308 natural antimicrobial compounds. Front Microbiol. 2012;3:287.
50. Demirel Z, Yilmaz-Koz FF, Karabay-Yavasoglu UN, Ozdemir G, Sukatar A. Antimicrobial and antioxidant activity of brown algae from the Aegean Sea. J Serb Chem Soc. 2009;74:619–28.
51. Cavallo RA, Acquaviva MI, Stabili L, Cecere E, Petrocelli A, Narracci M. Antibacterial activity of marine macroalgae against fish pathogenic Vibrio species. Cent Eur J Biol. 2013;8:646–53.
52. Bhagavathy S, Sumathi P, Jancy Sherene Bell I. Green algae Chlorococcum humicola a new source of bioactive compounds with antimicrobial activity. Asian Pac J Trop Biomed. 2011;1:S1–S7.
53. Manilal A, Sujith S, Kiran GS, Selvin J, Shakir C, Gandhimathi R, Lipton AP Antimicrobial potential and seasonality of red algae collected from the southwest coast of India tested against shrimp, human and phytopathogens. Ann Microbiol. 2009;59:207–19.
54. Bala N, Aitken EA, Cusack A, Steadman KJ. Antimicrobial potential of australian macrofungi extracts against foodborne and other pathogens. Phytother Res. 2012;26(3):465–9.
55. Kitzberger CSG, Smânia Jr A, Pedrosa RC, Ferreira SR. Antioxidant and antimicrobial activities of shiitake (Lentinula edodes) extracts obtained by organic solvents and supercritical fluids. J Food Eng. 2007;80:631–8.
56. Barros L, Calhelha RC, Vaz JA, Ferreira IC, Baptista P, Estevinho LM. Antimicrobial activity and bioactive compounds of Portuguese wild edible mushrooms methanolic extracts. Eur Food Res Technol. 2007;225:151–6.
57. Öztürk M, Duru ME, Kivrak ?, Mercan-Do?an N, Türkoglu A, Özler MA. In vitro antioxidant, anticholinesterase and antimicrobial activity studies on three Agaricus species with fatty acid compositions and iron contents: A comparative study on the three most edible mushrooms. Food Chem Toxicol. 2001;49:1353–60.
58. Tajkarimi M, Ibrahim S, Cliver D. Antimicrobial herb and spice compounds in food. Food Control. 2010;21(9):1199–218.
59. Hayek SA, Gyawali R, Ibrahim SA. Antimicrobial natural products. In: Méndez-Vilas A Editor. Microbial pathogens and strategies for combating them: science, technology and education. Norristown, PA: Formatex Research Center. 2013;V(2), pp. 910–21.
60. Savoia D. Plant-derived antimicrobial compounds: alternatives to antibiotics. Future Microbiol. 2012;7(8):979–90.
61. Negi PS. Plant extracts for the control of bacterial growth: efficacy, stability and safety issues for food application. Int J Food Microbiol. 2012;156(1);7–17.
62. Ciocan ID, B?ra I. Plant products as an-timicrobial agents. Universitatii ale ?tiin?ifice Analele Alexandru Ioan Cuza. 2007; Tom VIII.
63. Lai P, Roy J. Antimicrobial and chemopreventive properties of herbs and spices. Curr Med Chem. 2004;11(11):1451–60.
64. Ultee A, Bennik MHJ, MoezelaarR. The phenolic hydroxyl group of carvacrol is essential for action against the foodborne pathogen Bacillus cereus. Applied and environmental microbiology. 2002;68,(4):1561-1568.
65. Dorman HJ, Deans SG. Antimicrobial agents from plants: antibacterial 1110 activity of plant volatile oils. J Appl Microbiol. 2000;88(2):308–16.
66. Balasundram N, Sundram K, Samman S. Phenolic compounds in plants and agri1003 industrial by-products: antioxidant activity, occurrence, and potential uses. Food Chem. 2006;99(1):191–203.
67. Engels C, Knödler M, Zhao YY, Carle R, Gänzle MG, Schieber A. Antimicrobial activity of gallotannins isolated from mango (Mangifera indica L.) kernels. J Agric Food Chem. 2009;57(17):7712–8.
68. Figuerola F, Hurtado MA, Estévez AMA, Chiffelle I, Asenjo F. Fibre concentrates from apple pomace and citrus peel as potential fibre sources for food enrichment. Food Chem. 2005;91(3):395–401.
69. Al-Zoreky N. Antimicrobial activity of pomegranate (Punica granatum L.) fruit peels. Int J Food Microbiol. 2009;134(3):244–8.
70. Negi P, Jayaprakasha G. Antioxidant and antibacterial activities of Punica granatum peel extracts. J Food Sci. 2003;68(4):1473–7.
71. Machado T, Pinto A, Pinto M, Leal I, Silva M, Amaral A, et al. In vitro activity of Brazilian medicinal plants, naturally occurring naphthoquinones and their analogues, against methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2003;21(3):279–84.
72. Mandalari G, Bennett R, Bisignano G, Trombetta D, Saija A, Faulds C, et al. Antimicrobial activity of flavonoids extracted from bergamot (Citrus bergamia Risso) peel, a byproduct of the essential oil industry. J Appl Microbiol. 2007;103(6):2056–64.
73. Fattouch S, Caboni P, Coroneo V, Tuberoso CI, Angioni A, Dessi S, et al. Antimicrobial activity of Tunisian quince (Cydonia oblonga Miller) pulp and peel polyphenolic extracts. J Agric Food Chem. 2007;55(3):963–9.
74. Abdalla AE, Darwish SM, Ayad EH, El-Hamahmy RM. Egyptian mango by product 2: Antioxidant and antimicrobial activities of extract and oil from mango seed kernel. Food Chem. 2007;103(4):1141–52.
75. Kabuki T, Nakajima H, Arai M, Ueda S, Kuwabara Y, Dosako SI. Characterization of novel antimicrobial compounds from mango (Mangifera indica L.) kernel seeds. Food Chem. 2000;71(1):61–6.
76. Kanatt SR, Arjun K, Sharma A. Antioxidant and antimicrobial activity of legume hulls. Food Res Int. 2011;44(10):3182–7.
77. Adebowale B, Ogunjobi M, Olubamiwa O, Olusola-Taiwo M, Omidiran V. Quality improvement and value addition of processed fish (Clarias gariepinus) using phenolic compounds in coffee pulp smoke. Int Res J Agric Sci Soil Sci. 2012;2(13):520–4.
78. Lönnerdal, B. Biological effects of novel bovine milk fractions. Nestle Nutr Workshop Ser Pediatr Program. 2011;67:41–54.
79. USDA-FSIS. Safe and suitable ingredients used in the production of meat, poultry, and egg products. FSIS Dir. 7120.1 Revision 2. Annapolis, MA: USDA-FSIS; 2010.
80. Al-Nabulsi AA, Holley RA. Effect of bovine lactoferrin against Carnobacterium viridans. Food Microbiol. 2005;22(2):179–87.
81. Tikhonov VE, Stepnova EA, Babak VG, Yamskov IA, Palma-Guerrero J, Jansson HB, et al. Bactericidal and antifungal activities of a low molecular weight chitosan and its N-/2(3)-(dodec-2-enyl) succinoyl/-derivatives. Carbohydr Polym. 2006;64(1):66–72.
82. Chung YC, Yeh JY, Tsai CF. Antibacterial characteristics and activity of water-soluble chitosan derivatives prepared by the Maillard reaction. Molecules. 2011;16(10):8504–14.
83. Cegielska-Radziejewska R, Lesnierowski G, Kijowski J. Antibacterial activity of hen egg white lysozyme modified by thermochemical technique. Eur Food Res Technol. 2009;228(5):841–5.
84. Suthiluk S, Kamhangwong D, Benjakul S. Antimicrobial activity of some potential active compounds against food spoilage microorganisms. Afr J Biotechnol. 2012;11(74):13914–21.
85. Schanbacher F, Talhouk R, Murray F, Gherman L, Willett L. Milk-borne bioactive peptides. Int Dairy J. 1998;8(5–6):393–403.
86. McCann K, Shiell B, Michalski W, Lee A, Wan J, Roginski H, et al. Isolation and characterisation of a novel antibacterial peptide from bovine αS1-casein. Int Dairy J. 2006;16(4):316–23.
87. Szwajkowska M, Wolanciuk A, Bar?owska J, Król J, Litwiñczuk Z. Bovine milk proteins as the source of bioactive peptides influencing the consumers’ immune system–a review. Anim Sci Pap Rep. 2011;29(4):269–80.
88. Arqués JL, Fernández J, Gaya P, Nuñez M, Rodríguez E, Medina M. Antimicrobial activity of reuterin in combination with nisin against food-borne pathogens. Int J Food Microbiol. 2004;95(2):225–9.
89. Rajendran K, Nagappan R, Ramamurthy K. Short Communication A study on the bactericidal effect of nisin purified from Lactococcus lactis. Ethiop J Biol Sci. 2013;10:1.
90. Arqués JL, Rodríguez E, Nuñez M, Medina M. Combined effect of reuterin and lactic acid bacteria bacteriocins on the inactivation of food-borne pathogens in milk. Food Control. 2011;22(3):457–61.
91. Bian L, Molan AL, Maddox I, Shu Q. Antimicrobial activity of Lactobacillus reuteri DPC16 supernatants against selected food borne pathogens. World J Microbiol Biotechnol. 2011;27(4):991–8.
92. Willis WL, King K, Iskhuemhen OS, Ibrahim SA. Administration of mushroom extract to broiler chickens for bifidobacteria enhancement and Salmonella reduction. J Appl Poult Res. 2009;18(4):658–64.
93. Guedes AC, Barbosa CR, Amaro HM, Pereira CI, Malcata FX. Microalgal and cyanobacterial cell extracts for use as natural antibacterial additives against food pathogens. Int J Food Sci Technol. 2011;46(4):862–6.
94. Kalyoncu F, Oskay M, Sa?lam H, Erdo?an TF, Tamer AÜ. Antimicrobial and antioxidant activities of mycelia of 10 wild mushroom species. J Med Food. 2010;13(2):415–9.
95. Ramesh C, Pattar MG. Antimicrobial properties, antioxidant activity and bioactive compounds from six wild edible mushrooms of western ghats of Karnataka, India. Phytother Res. 2010;2(2):107.
96. Sozer N, Kokini JL. Nanotechnology and its applications in the food sector. Trends Biotechnol. 2009;27(2):82–9.
97. Zou Y, Lee HY, Seo YC, Ahn J. Enhanced antimicrobial activity of nisin-loaded liposomal nanoparticles against Foodborne Pathogens. J Food Sci. 2012;77(3):M165–70.
98. Prombutara P, Kulwatthanasal Y, Supaka N, Sramala I, Chareonpornwattana S. Production of nisin-loaded solid lipid nanoparticles for sustained antimicrobial activity. Food Control. 2012;24(1):184–90.
99. Tripathi KD. Textbook of essentials of medical pharmacology. 8th ed. Noida: Jaypee; 2019.
100. O’Neill J. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance. London: HM Government and Wellcome Trust; 2016.
101. World Health Organization, Regional office for South-East Asia. Jaipur declaration on antimicrobial resistance. Geneva: WHO; 2011.
102. Department of Health, Department for Environment Food and Rural Affairs. UK Five Year Antimicrobial Resistance Strategy 2013 to 2018. London: Government of UK; 2013.
103. Government of India. National Action Plan on Antimicrobial Resistance (NAP-AMR) 2017–2021. New Delhi: Government of India; 2017.
104. Rrang and Dales. Textbook of pharmacology. 8th ed. Amsterdam: Elsevier; 2015.
105. Taneja N, Sharma M. Antimicrobial resistance in the environment: The Indian scenario. Indian J Med Res. 2019;149(2):119–28.
106. Kahn LH. Antimicrobial resistance: a one health perspective. Trans R Soc Trop Med Hyg. 2017;111:255–60.
107. Gandra S, Joshi J, Trett A, Lamkang A, Laxminarayan R. Scoping report on antimicrobial resistance in India. Washington, DC: Center for Disease Dynamics, Economics and Policy; 2017. Available from: http://www.dbtindia.nic.in/wp-content/uploads/ScopingreportonAntimicrobialresistanceinIndia.pdf (accessed April 15, 2017).
108. Chaudhry D, Tomar P. Antimicrobial resistance: the next big pandemic. Int J Community Med Public Health. 2017;4:2632–6.
109. Swaminathan S, Prasad J, Dhariwal AC, Guleria R, Misra MC, Malhotra R, et al. Strengthening infection prevention and control and systematic surveillance of healthcare associated infections in India. BMJ. 2017;358:j3768.
110. Laxminarayan R, Matsoso P, Pant S, Brower C, Røttingen JA, Klugman K, et al. Access to effective antimicrobials: A worldwide challenge. Lancet. 2016;387:168–75.
111. Morgan DJ, Okeke IN, Laxminarayan R, Perencevich EN, Weisenberg S. Non-prescription antimicrobial use worldwide: a systematic review. Lancet Infect Dis. 2011;11:692–701.
112. Lundborg CS, Tamhankar AJ. Antibiotic residues in the environment of South East Asia. BMJ. 2017;358:j2440.
113. Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509:612–6.
114. Chereau F, Opatowski L, Tourdjman M, Vong S. Risk assessment for antibiotic resistance in South East Asia. BMJ. 2017;358:j3393.
115. Singer AC, Shaw H, Rhodes V, Hart A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front Microbiol. 2016;7:1728.
116. Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global antibiotic consumption 2000 to 2010: an analysis of national pharmaceutical sales data. Lancet Infect Dis. 2014;14:742–50.
117. Ambesh P, Ambesh SP. Open defecation in India: a major health hazard and hurdle in infection control. J Clin Diagn Res. 2016;10:IL01–2.
118. Bain R, Cronk R, Hossain R, Bonjour S, Onda K, Wright J, et al. Global assessment of exposure to faecal contamination through drinking water based on a systematic review. Trop Med Int Health. 2014;19:917–27.
119. Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A. 2015;112:5649–54.
120. Hao H, Cheng G, Iqbal Z, Ai X, Hussain HI, Huang L, et al. Benefits and risks of antimicrobial use in food-producing animals. Front Microbiol. 2014;5:288.
121. Sahu R, Saxena P. Antibiotics in chicken meat. PML/PR-48/2014. New Delhi, India: Centre for Science and Environment, Centre for Science and Environment, India; 2014. Available from: https://cdn.cseindia.org/userfiles/Antibiotics%20in%20Chicken_Lab%20Report_Final%2029%20July.pdf (accessed April 15, 2017).
122. Goutard FL, Bordier M, Calba C, Erlacher-Vindel E, Góchez D, de Balogh K, et al. Antimicrobial policy interventions in food animal production in South East Asia. BMJ. 2017;358:j3544.
123. Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DG. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics. 2015;16:964.
124. Rehman MS, Rashid N, Ashfaq M, Saif A, Ahmad N, Han JI, et al. Global risk of pharmaceutical contamination from highly populated developing countries. Chemosphere. 2015;138:1045–55.
125. Lata P, Ram S, Shanker R. Multiplex PCR based genotypic characterization of pathogenic vancomycin resistant Enterococcus faecalis recovered from an Indian river along a city landscape. Springerplus. 2016;5:1199.
126. Mutiyar PK, Mittal AK. Risk assessment of antibiotic residues in different water matrices in India: key issues and challenges. Environ Sci Pollut Res. 2014;21:7723–36.
127. Diwan V, Tamhankar AJ, Khandal RK, Sen S, Aggarwal M, Marothi Y, et al. Antibiotics and antibiotic-resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health. 2010;10:414.
128. Duong HA, Pham NH, Nguyen HT, Hoang TT, Pham HV, Pham VC, et al. Occurrence, fate and antibiotic resistance of fluoroquinolone antibacterials in hospital wastewaters in Hanoi, Vietnam. Chemosphere. 2008;72:968–73.
129. Wichmann F, Udikovic-Kolic N, Andrew S, Handelsman J. Diverse antibiotic resistance genes in dairy cow manure. MBio. 2014;5:e01017.
130. Clarke BO, Smith SR. Review of ‘emerging’ organic contaminants in biosolids and assessment of international research priorities for the agricultural use of biosolids. Environ Int. 2011;37:226–47.
131. Henriksson PJ, Troell M, Rico A. Antimicrobial use in aquaculture: some complementing facts. Proc Natl Acad Sci U S A. 2015;112:E3317.
132. World Health Organization. Antimicrobial resistance: global report on surveillance. Geneva: WHO; 2014.
133. Government of India. National Action Plan on Antimicrobial Resistance (NAP-AMR). New Delhi: Government of India; 2017.
134. Ministry of Health and Family Welfare, Government of India. National Health Policy. New Delhi: MOHFW; 2017.
135. Government of India. Food Safety and Standards Authority of India. Annual Report 2017. New Delhi: Ministry of Health and Family Welfare, Government of India; 2017.
136. Tyurin AP, Alferova VA, Paramonov AS, Shuvalov MV, Kudryakova GK, Rogozhin EA, et al. Gausemycins A,B: cyclic lipoglycopeptides from Streptomyces sp. Angew Chem Int Ed. 2021;60:18694–703.
137. Monciardini P, Iorio M, Maffioli S, Sosio M, Donadio S. Discovering new bioactive molecules from microbial sources. Microb Biotechnol. 2014;7:209–20.
138. Boruta T. A Bioprocess perspective on the production of secondary metabolites by Streptomyces in submerged co-cultures. World J Microbiol Biotechnol. 2021;37:171.
139. Arora D, Gupta P, Jaglan S, Roullier C, Grovel O, Bertrand S. Expanding the chemical diversity through microorganisms co-culture: current status and outlook. Biotechnol Adv. 2020;40:107521.
140. Caudal F, Tapissier-Bontemps N, Edrada-Ebel RA. Impact of co-culture on the metabolism of marine microorganisms. Mar Drugs. 2022;20:153.
141. Peng XY, Wu JT, Shao CL, Li ZY, Chen M, Wang CY. Co-culture: Stimulate the metabolic potential and explore the molecular diversity of natural products from microorganisms. Mar Life Sci Technol. 2021;3:363–74.
142. Jung D, Liu L, He S. Application of in situ cultivation in marine microbial resource mining. Mar Life Sci Technol. 2021;3:148–61.
143. Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–9.
144. Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, et al. Use of ICHIP for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol. 2010;76:2445–50.
145. Gavrish E, Bollmann A, Epstein S, Lewis K. A Trap for in situ cultivation of filamentous actinobacteria. J Microbiol Meth. 2008;72:257–62.
146. Ben-Dov E, Kramarsky-Winter E, Kushmaro A. An in situ method for cultivating microorganisms using a double encapsulation technique: In situ method for cultivating microorganisms. FEMS Microbiol Ecol. 2009;68:363–71.
147. Jung D, Liu B, He X, Owen JS, Liu L, Yuan Y, et al. Accessing previously uncultured marine microbial resources by a combination of alternative cultivation methods. Microb Biotechnol. 2021;14:1148–58.
148. Davidson SL, Niepa THR. Micro-technologies for assessing microbial dynamics in controlled environments. Front Microbiol. 2022;12:745835.
149. Pope E, Cartmell C, Haltli B, Ahmadi A, Kerr RG. Microencapsulation and in situ incubation methodology for the cultivation of marine bacteria. Front Microbiol. 2022;13:958660.
150. Terekhov SS, Eliseev IE, Ovchinnikova LA, Kabilov MR, Prjibelski AD, Tupikin AE, et al. Liquid drop of DNA libraries reveals total genome information. Proc. Natl. Acad. Sci. USA 2020;117:27300–6.
151. Wollein Waldetoft K, Brown SP. Evolving antibiotic spectrum. Proc Natl Acad Sci USA. 2022;119:e2214267119.
152. Johnston CW, Badran AH. Natural and engineered precision antibiotics in the context of resistance. Curr Opin Chem Biol. 2022;69:102160.
153. Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–43 .
154. Avis T, Wilson FX, Khan N, Mason CS, Powell DJ. Targeted microbiome-sparing antibiotics. Drug Discov Today. 2021;26:2198–03.
155. Schorn MA, Verhoeven S, Ridder L, Huber F, Acharya DD, Aksenov AA, et al. A community resource for paired genomic and metabolomic data mining. Nat Chem Biol. 2021;17:363–8.
156. Louwen JJ, Medema MH, van der Hooft JJ. Enhanced correlation-based linking of biosynthetic gene clusters to their metabolic products through chemical cass matching. Microbiome 2023;11:13.
157. Muller E, Algavi YM, Borenstein E. The gut microbiome-metabolome dataset collection: A curated resource for integrative meta-analysis. NPJ Biofilms Microbiom. 2022;8:79.
158. Hou P, Nowak VV, Taylor CJ, Calcott MJ, Knight A, Owen JG. A genomic survey of the natural product biosynthetic potential of actinomycetes isolated from New Zealand lichens. mSystems. 2023;8:e01030-22.
159. Tenebro CP, Trono DJVL, Balida LAP, Bayog LKA, Bruna JR, Sabido EM, et al. Synergy between genome mining, metabolomics, and bioinformatics uncovers antibacterial chlorinated carbazole alkaloids and their biosynthetic gene cluster from Streptomyces tubbatahanensis sp. Nov., a novel actinomycete isolated from Sulu Sea, Philippines. Microbiol Spectr. 2023;11:e03661-22.
160. Milshteyn A, Colosimo DA, Brady SF. Accessing bioactive natural products from the human microbiome. Cell Host Microbe. 2018;23:725–36.
161. Chiumento S, Roblin C, Kieffer-Jaquinod S, Tachon S, Leprètre C, Basset C, et al. Ruminococcin C, a promising antibiotic produced by a human gut symbiont. Sci Adv. 2019;5:eaaw9969.
162. Zhang Q, Ren JW, Wang W, Zhai J, Yang J, Liu N, et al. A versatile transcription–translation in one approach for activation of cryptic biosynthetic gene clusters. ACS Chem Biol. 2020;15:2551–7.
163. Yoshimura A, Covington BC, Gallant É, Zhang C, Li A, Seyedsayamdost MR. Unlocking cryptic metabolites with mass spectrometry-guided transposon mutant selection. ACS Chem Biol. 2020;15:2766–74.
164. Covington BC, Seyedsayamdost MR. Guidelines for metabolomics-guided transposon mutagenesis for microbial natural product discovery. Methods Enzymol. 2022;66:305–23.
165. Hudson MA, Lockless SW. Elucidating the mechanisms of action of antimicrobial agents. mBio. 2022;13:e02240-21.
166. Rütten A, Kirchner T, Musiol-Kroll EM. Overview on strategies and assays for antibiotic discovery. Pharmaceuticals. 2022;15:1302.
167. Sergiev PV, Osterman IA, Golovina AY, Andreyanova ES, Laptev IG, Pletnev PI, et al. Application of reporter strains for screening of new antibiotics. Biochem Moscow Suppl Ser B. 2016;10:293–9.