INTRODUCTION
A cell-based assay for adipogenesis and adipocyte differentiation has become a powerful tool for in-depth studies on adipocytes related to the study of obesity. Most commonly, murine preadipocyte cell lines are used due to their ease of maintenance and passage. Research using cell and animal models has provided more comprehensive insights into metabolic pathways in obesity compared to human models, which are practically limited by their passage capacity and physiological properties. Over the past 5 years, more than 400 articles have utilized several different murine preadipocyte cell lines to study adipogenesis and obesity processes. These articles primarily focused on modifications of adipocyte differentiation methodologies and adjustments to the adipogenesis agent mixtures to suit different cell types. Although several adipogenesis induction protocols have been clearly delineated step-by-step, achieving consistent and complete success remains elusive. Various cell-based models are employed in obesity mechanism studies, such as 3T3-L1, 3T3-F442A, C3H10T1/2, and mouse stromal cells (OP9) [1-3]. All these models require precise protocols, specific inducing factors, and incubation times ranging from 3 to 18 days. The most well-known pre-adipocyte cell line, 3T3-L1, also faces challenges such as a lengthy 10-day to 2-week induction process, passaging difficulties, and culture condition limitations, often resulting in uncertain outcomes.
Most lipid accumulation and adipocyte differentiation assays have been evaluated using various biochemical techniques such as colorimetric quantification and flow cytometry. Although these techniques offer convenience, time efficiency, and reliable results, they also require a substantial laboratory budget. Additionally, each technique necessitates different chemicals, instruments, and usage criteria, and comes with its own set of limitations and challenges. To circumvent the complexities associated with biochemical methods, image-based analysis has been introduced and has become a significant field in biomedical research [4]. Mori et al. [5] developed a modified technique using a colorimetric assay combined with image analysis to quantify the total intracellular lipid content from an entire culture plate of induced pluripotent stem cells. Our previous study also employed image-based analysis to evaluate the anti-adipogenesis effects of plant extracts on L929 cells, yielding acceptable outcomes although direct comparisons to 3T3-L1 cells were not undertaken [6].
The murine fibroblast cell line, L929, has been employed in various cellular mechanism studies, including adipocyte differentiation and cytotoxicity tests before application in 3T3-L1 [7,8]. L929 cells can differentiate into pre-adipocytes and mature adipocytes without the need for inducing agents or prolonged incubation periods. However, there are few reports on the potential application of L929 as a cell-based model for studying adipogenesis mechanisms. Spontaneous differentiation into pre-adipocytes can be achieved by maintaining L929 cells at 70%–80% confluence for 5 days, during which lipid droplets accumulate in the cytoplasm [6]. The results indicate that L929 could serve as an alternative cell model for adipogenesis and obesity studies.
Previous studies on anti-adipogenesis have demonstrated that (-)-epigallocatechin gallate (EGCG), a phenolic bioactive molecule abundant in green tea, effectively reduces adipogenesis and fat accumulation in various cell types [9]. Appropriate concentrations of EGCG have been shown to inhibit the expression of adipogenic regulatory genes and lipogenic enzymes in murine preadipocytes [10]. Significant reductions in body weight, fat tissue weight, and plasma lipid levels were observed in obese mice supplemented with EGCG [11]. The inhibition of adipogenesis, adipocyte differentiation, and triglyceride accumulation by EGCG has been documented in multiple cell lines, including 3T3-L1 [9]. However, no research has yet reported the effects of EGCG on the inhibition of adipogenesis in L929 cells.
L929 and NIH3T3 are both fibroblast cell lines used in research, but they have different properties and behaviors. The spontaneous accumulation of fat in L929 and NIH3T3 cells has been reported [12]. The choice between these cell lines for spontaneous adipocyte differentiation can be influenced by various factors. The potential advantages of using L929 cells over NIH3T3 cells for spontaneous adipocyte differentiation include differentiation efficiency. L929 cells might have a higher propensity for adipocyte differentiation under specific conditions compared to NIH3T3 cells. If the L929 cells differentiate more readily or consistently, this could make them a more reliable choice for studies focusing on adipogenesis. L929 cells may also show greater lipid accumulation when differentiating into adipocytes. This can be advantageous for experiments where measuring lipid content or observing lipid droplet formation is a key objective. The gene expression profiles related to adipogenesis might also differ between the two cell lines. L929 cells could express certain adipogenic markers more robustly, making them a better model for studying the molecular mechanisms of adipocyte differentiation. L929 cells have different growth rates and requirements compared to NIH3T3 cells. Depending on the experimental design, the growth characteristics of L929 cells could be more favorable, such as faster growth rates or lower nutrient requirements [12]. L929 cells might respond more effectively to adipogenic induction agents, such as certain hormones or chemicals. This responsiveness could lead to more efficient and reproducible differentiation and utilize less xenobiotic in the experiments. L929 cells might also be more amenable to certain experimental techniques or assays, such as staining, imaging, or gene knockdown/knockout experiments.
This study represents the first investigation into the effects of EGCG on the anti-adipogenesis of L929 cells using an image-based analysis technique. To provide a comprehensive comparison, the effects of EGCG on 3T3-L1 cells were also assessed concurrently using biochemical approaches. The study evaluated cellular sensitivity, anti-adipogenesis, and adipocyte-related gene expression in pre-adipocytes and mature adipocytes of both L929 and 3T3-L1 cell lines. The findings from this research provide a significant understanding of the potential use of L929 cells as a model for adipocyte differentiation. Additionally, the image-based assay offers a cost-effective, fast, and technologically accessible method, presenting a viable alternative for preliminary studies and start-up laboratories.
MATERIALS AND METHODS
Cell culture
The pre-adipocytes cell line, 3T3-L1 and L929 were purchased from (ATCC, Manassas, USA). The cells were cultured in a 25 cm2 tissue culture flask (Nunc©, Denmark), nourished with a complete medium (DMEM, 10% (v/v) FBS, 1X antibiotics-antimycotic), and maintained under a standard culture condition with 37°C, 95% relative humidity in 5% CO2 incubator. Trypsinization using 1% (v/v) trypsin-EDTA was performed every 2–3 days or before the cell reached 70% confluency to avoid differentiation and detachment. All chemicals were purchased from Thermo Fisher Scientific, USA.
Cellular sensitivity assay
The commercial polyphenols, (-)-epigallocatechin gallate (EGCG, C22H18O11, MW = 458.37 g/mol, E4143, Merk, Germany), was dissolved in absolute ethanol (Sigma-Aldrich, USA) to prepare a stock solution. Working concentrations of EGCG, at 10, 50, and 100 µM were freshly prepared and directly diluted in a complete medium. The 50,000 cell/ml of L929 or 30,000 cell/ml of 3T3-L1 were seeded onto the 96-well plate for 24 hours in advance. To perform the test, the medium in the culture plate was gently discarded, varied concentrations of EGCG were added, and incubated the cells in a standard culture condition for 72 hours. Complete medium without EGCG and 0.1% (v/v) dimethylsulfoxide were assigned as a control. Five replicates were prepared for each treatment. Cell sensitivity relating to viability was evaluated by MTT-formazan colorimetric assay [13]. The absorbance at 570 nm (A570) was read out using a microplate-reader (Rayto TR-2100C, Shenzhen, China). The A570 value was then calculated to % relative viability in comparison to the control by using the equation (1) as shown below. PriProbit Program ver. 1.63 [14] was employed to estimate the IC50 of EGCG which was further applied in the anti-adipogenesis experiment.
when A570 represented as the absorbance value
Adipogenic induction
The culture surface for adipocyte differentiation must be a pre-coated culture surface with 0.1% (v/v) bovine skin gelatin (type B) for 30 minutes at 37°C (a standard plate coating protocol as described by ATCC® (PCS-999-027™). After that, the gelatin solution was discarded, and the culture surface was gently washed once with PBS before the addition of the complete medium onto the culture plate. This gelatin-coated plate is kept at standard culture conditions for 3 days. Before cell seeding, the complete medium in the plate was discarded and the culture surface was ready to use.
Spontaneous induction in L929
The optimal condition for L929 spontaneous adipogenic differentiation was determined previously [4]. The single-cell suspension of trypsinized-L929 at 50,000 cell/ml was seeded onto the gelatin-coated plate and maintained under a standard culture condition for 5 days. The fresh medium was renewed every 2 days.
Chemical induction in 3T3-L1
This experiment was performed as described [15]. In brief, 3T3-L1 at 30,000 cell/ml were seeded in a culture plate for 2 days and then incubated with the sequential-inductive medium (a completed media + 0.5 mM IBMX (3-isobutyl-1-methylxanthine) + 1 mM DEX (dexamethasone) + 167 nM insulin) for 48 hours. After that, the cells were maintained in the insulin medium (a completed medium + 167 nM insulin) for 14 days to obtain mature adipocytes.
After the adipogenesis induction processes were completely undertaken, the cells were fully differentiated to mature adipocytes of L929 and 3T3-L1.
Lipid staining and quantification
Oil Red O staining
Lipid droplet accumulating in the cells was stained and quantified by Oil Red O soluble dye as previously described [16]. Cells were gently washed with PBS before fixing in 10% (w/v) formaldehyde for an hour. The fixed cell was thoroughly rinsed with PBS twice, incubated with 0.3% (w/v) Oil Red O (Sigma-Aldrich, USA) in 60% (v/v) isopropanol (Sigma-Aldrich, USA), 200 µl per well for 15 minutes and rinsed 3–4 times with distilled water. At this step, the lipid droplets in the cells were turned to vivid red colors which could be observed under the microscope.
Image-based lipid droplet quantification in L929
Oil Red O and methylene blue (MB) counterstaining
The L929 containing lipid droplets was first stained by Oil Red O as described above before counterstain with MB adapted from [17]. After Oil Red O staining was completed by rinsing with distilled water, 0.1% (w/v) MB solution (Sigma-Aldrich, USA) was wisely dropped onto the cells. Incubated at room temperature for 1–3 minutes before gently rinsing with distilled water twice, or until completely removing the excessed dye. The stained cells could be observed under the microscope and ready to quantify the lipid droplets by the image-based assay.
Lipid droplet area measurement
The stained cells were randomly photomicrographed by a CCD camera installed in the microscope (all microscopic instruments were from Olympus, Japan). The images were taken from each treatment for five random areas, with at least 200 cells per area, approximately 105 cells per treatment in total. The image-based analysis was performed through the captured images in *.TIFF format and further processed by ImageJ software (https://imagej.nih.gov) as described [18]. Raw photomicrographs or pre-processing images were adjusted color thresholds to enhance the intensity of red color of lipid droplets. The measurement areas were converted from micrometers to pixels before image processing. The function of the thresholding method and color was set by selecting commands on the menu bar of ImageJ as “Image”, “Adjust”, and then “Color Threshold”. A new window of the menu was shown as “Threshold Color (experimental)”, then selected from the drop-down menu as “MaxEntropy” and “Red”, respectively, resulting in the established post-processing images. The post-processing images exhibited vibrant red color lipid droplets and blue areas of the cells. Only the areas of red-lipid droplets were measured by selecting the command “Analyze” followed by “Analyze Particles” on the program bar. A new window of “Analyze Particles” popped up, and the unit of measurement particle was shown in zero to infinity µm2. The “Counter Masks” and “Summarize” on drop-down menus were selected to confirm the area of measurement, then clicked “OK”. The results were provided in areas of lipid droplets per cells in µm2 of each picture. At least 200 cells were collected and evaluated in the lipid droplet area. The % relative lipid amount from each treatment was then calculated in comparison to a control by using equation (2) as shown below.
when total lipid and cell area represented in µm2
Colorimetric lipid quantification in 3T3-L1
First, the cells were stained with Oil Red O as mentioned above. To quantify the amount of lipid droplets, Oil Red O dye was extracted from the cell by adding 40 µl of 50% (v/v) isopropanol into each well. Then the absorbance at 510 nm (A510) was measured using the microplate reader. The % relative lipid amount was calculated in comparison to a control (insulin-medium without EGCG) by using the equation (3) as shown below.
when A510 represented as the absorbance value.
Effects of EGCG on anti-adipogenesis
L929
The 50,000 cells/ml of L929 were plated on the gelatin-coat surface and maintained in the standard culture condition for 24 hours. The varied concentration of EGCG was prepared to 10, 20, and 40 µM (referred to the cellular sensitivity assay and IC50). The cells were exposed to the complete medium containing varied EGCG concentrations for 3 days. The complete medium without EGCG (0 µM) was set as treatment control. The lipid droplet quantification from each treatment was measured by the image-based assay as described above.
3T3-L1
For 3T3-L1, the anti-adipogenesis evaluation was carried on during the 14 days of adipogenesis induction (as mentioned above). First, the 30,000 cells/ml of 3T3-L1 were plated on the gelatin-coat surface and nourished with the sequential-inductive medium for 2 days. Thereafter, the insulin medium containing varying concentrations of EGCG was prepared to reach a final concentration at 10, 50, and 100 µM (referred to the cellular sensitivity assay and IC50). The cells were maintained in the insulin-EGCG medium for 14 days. The fresh medium was replaced every 2 days. The insulin medium without EGCG (0 µM) was set as treatment control. All treatment was performed in five replicates. The lipid droplet quantification was measured by Oil Red O colorimetric assay as mentioned above.
Viability test
Both pre-adipocyte L929 and 3T3-L1 were prepared and exposed to the EGCG at varying concentrations with the defined incubation time. All treatments and controls were set as indicated in those topics. Cell viability was evaluated by MTT-formazan colorimetric assay as described above (see “Cellular sensitivity assay”).
Adipogenesis-related gene expression analysis
The cells were primarily prepared and exposed to 10 µM EGCG for 3 days (L929) or 100 µM EGCG for 14 days (3T3-L1). Either L929 or 3T3-L1 mature adipocytes were harvested for RNA collecting using the Nucleospin® RNA isolation kit (Macherey-Nagel, Germany). Total mRNA was used as a template for cDNA synthesis by ReverTra Ace® qPCR RT master mix (Toyobo Co., Japan) as mentioned in the manufacturer’s protocol. The related gene, peroxisome proliferator-activated receptor-gamma (PPARγ), CCAAT enhancer-binding protein-alpha (C/EBPa), adipocyte protein 2 (aP2), and preadipocyte factor-1 (Pref-1), were examined by quantitative real-time PCR (SensiFASTTM SYBER® No-ROX Kit (Bioline, UK) and operated under the EcoTM Real-Time PCR System (Illumina, Inc., USA). All reactions were set up for five replicates individually. The β-actin was assigned as a housekeeping gene (an internal control). Primer sequences (Table 1) were designed by Primer 3, followed by specificity verification through BLAST tools. The PCR reaction mixture was first denaturated at 94°C for 3 minutes followed by 94°C for 3 seconds (40 cycles) and terminated at 72°C for 30 seconds. The CT values were normalized, and the calculated gene expression level used the 2-ΔΔCT method [19]. The expression of each gene was compared to none-exposed mature adipocytes (controls of L929 or 3T3-L1) and then presented as a histogram of relative expressions.
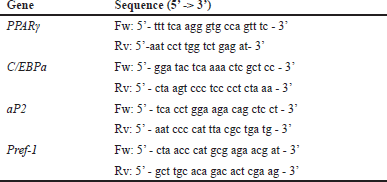 | Table 1. Primer sequences. [Click here to view] |
Statistical analysis
All data are represented in mean ± standard deviation (SD) from five replicates. Statistical difference was analyzed by one-way ANOVA and Duncan’s multiple range test at p ≤ 0.05.
RESULTS
Mature adipocyte morphology of L929 and 3T3-L1
After pre-adipocyte L929 and 3T3-L1 cells were plated onto a gelatin-coated surface (Fig. 1A, 1B), they were consistently maintained until confluency, where they progressively differentiated into mature adipocytes (MA) (Fig. 1C, 1D). Oil Red O staining vividly displayed the amount and shape of lipid droplets within the cells (Fig. 1E, 1F).
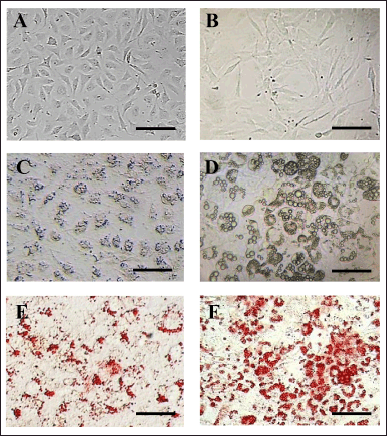 | Figure 1. Fresh pre-adipocytes of (A) L929 and (B) 3T3-L1 cells after plating onto a gelatin-coated surface for 24 hours. (C) Mature adipocytes (MA) of L929 and (D) MA-3T3-L1 cells showing lipid droplet accumulation. Oil Red O staining illustrates lipid accumulation in red, as observed in (E) MA-L929 and (F) MA-3T3-L1 cells. Scale bar = 150 µm. [Click here to view] |
24 hours after plating, pre-adipocyte L929 cells exhibited a spindle-like morphology with a prominent nucleus (Fig. 1A). These cells progressively differentiated, accumulated lipid droplets, and by day 5 of culture, fully displayed the characteristics of mature adipocytes (MA-L929). The cell bodies of MA-L929 became larger and more oval to round, with numerous tiny lipid droplets dispersed around the nucleus in the cytoplasm, which were challenging to identify without staining (Fig. 1C). Oil Red O staining made these lipid droplets clearly visible (Fig. 1E).
For pre-adipocyte 3T3-L1 cells, complete adherence to the surface and a spindle-like shape were observed after 24 hours of plating on the gelatin-coated plate (Fig. 1B). During the induction process, significant morphological changes in pre-adipocyte 3T3-L1 cells were noticeable under the microscope. By day 14, the mature adipocytes (MA-3T3-L1) exhibited a more oval shape and accumulated large lipid droplets in the cytoplasm (Fig. 1D), which were clearly visible with Oil Red O staining (Fig. 1F).
Cellular sensitivity to EGCG
The cellular sensitivity was evaluated using % viability after a 72-hour exposure to different dilutions of EGCG; 0, 10, 50, and 100 µM. L929 exhibited greater sensitivity to EGCG than 3T3-L1 demonstrating a decrease in % viability at a higher concentration of EGCG (p ≤ 0.05) (Fig. 2). Probit analysis calculation indicated the IC50 of L929 and 3T3-L1 which was 78.47 and 1.39 × 104 µM EGCG, respectively. The EGCG concentration ranking below IC50 was considered to apply in the anti-adipogenesis evaluation.
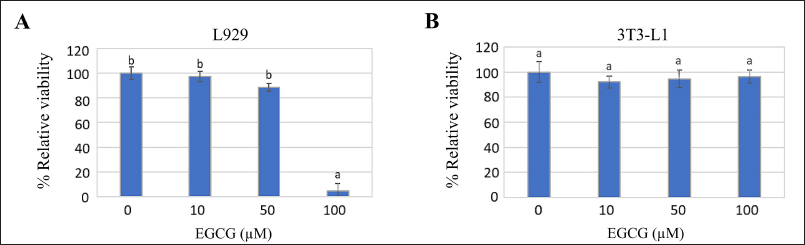 | Figure 2. Cellular sensitivity of (A) L929 and (B) 3T3-L1 cells, evaluated by assessing the percentage of relative viability after exposure to different concentrations of EGCG for 72 hours. The letters (a and b) above the bars indicate significant differences among the EGCG concentrations for each cell type (p ≤ 0.05). [Click here to view] |
Effects of EGCG on anti-adipogenesis
According to the EGCG sensitivity test of L929 (IC50 = 78.47 µM) and 3T3-L1 (IC50 = 1.39 × 104 µM), the concentrations below the IC50 of each cell were applied in this experiment to avoid cell death. EGCG at varying concentrations was exposed to the cells throughout the adipogenesis processing for 3 or 14 days for L929 or 3T3-L1, respectively.
Morphology and lipid accumulation
For L929 cells, after 3 days of EGCG exposure, the cells were fixed and stained with Oil Red O and MB. Pre-processing photomicrographs were adjusted using ImageJ software to obtain post-processing images that clearly exhibited contrasting red lipid droplets and blue cell bodies. The counter mask images displayed the actual areas of lipid droplet measurement (Fig. 3). The area of red lipid droplets was measured from the post-processed images. Quantitative data for each treatment were then calculated and represented as the % relative lipid amount in parallel with % relative viability.
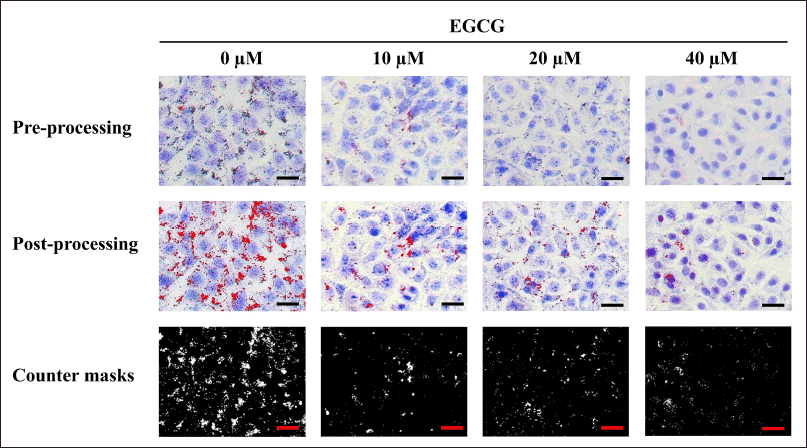 | Figure 3. The pre-processing photomicrographs of MA-L929 cells were adjusted using ImageJ software to produce post-processing images, facilitating the measurement of red lipid droplets and blue cytoplasmic cell areas. Scale bar = 100 µm. [Click here to view] |
For 3T3-L1 cells, they were exposed to EGCG for 14 days during adipogenic induction. The cells were subsequently photographed (Fig. 4), and the lipid content was biochemically quantified. The % relative lipid amount and viability were then verified and reported.
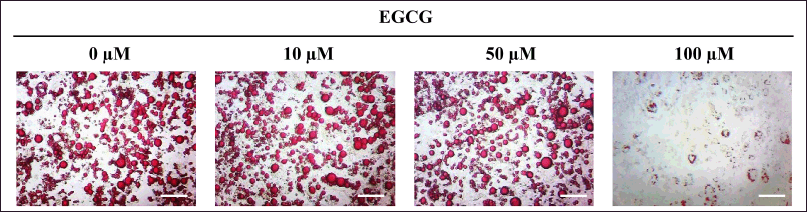 | Figure 4. Photomicrographs of lipid droplets accumulating in MA-3T3-L1 cells exposed to different concentrations of EGCG for 14 days during adipogenic induction. The amount of cytoplasmic lipid was biochemically quantified using a colorimetric assay. Scale bar = 100 µm. [Click here to view] |
The relative viability of MA-L929 and MA-3T3-L1 cells during the anti-adipogenesis experiment ranged from 94% to 97%, with no statistically significant difference from the controls (0 μM EGCG) (p ≤ 0.05) (Fig. 5). The percentage of lipid content in L929 cells, as measured by image-based assay, is shown in Figure 5A, while the results of colorimetric quantification for 3T3-L1 cells are presented in Figure 5B. Notable differences were observed between MA-L929 and MA-3T3-L1 in their responses to EGCG. In MA-L929 cells, 10 - 40 μM EGCG significantly reduced lipid content (p ≤ 0.05) (Fig. 5A), despite apparent reductions in lipid droplets at other concentrations visible in the photomicrographs (Fig. 3). Conversely, in MA-3T3-L1 cells, a significant reduction in lipid content was observed only at 100 μM EGCG (p ≤ 0.05) (Fig. 5B).
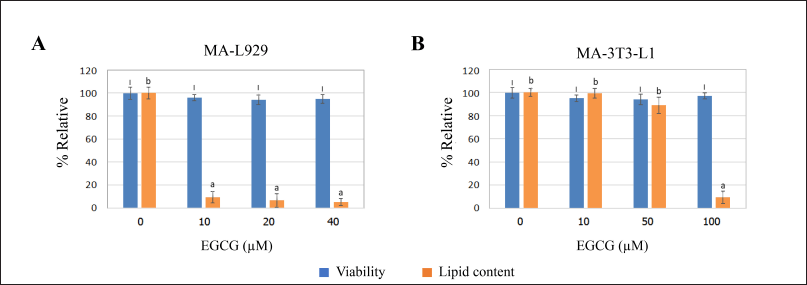 | Figure 5. Percent relative viability (assessed by the MTT assay) and lipid content influenced by EGCG at various concentrations were evaluated in (A) MA-L929 cells (3 days) and (B) MA-3T3-L1 cells (14 days) using image-based analysis and colorimetric Oil Red O staining, respectively. The letters (I) above the bars indicate significant differences in viability across EGCG concentrations, while (a and b) denote significant differences in lipid content within each cell type (p ≤ 0.05). [Click here to view] |
Gene expression patterns
Gene expression analysis was conducted on L929 and 3T3-L1 cells exposed to 10 µM EGCG for 3 days and 100 µM EGCG for 14 days, respectively. These EGCG concentrations were chosen based on their demonstrated anti-adipogenesis potential. The relative expression levels of adipogenesis-related genes were analyzed and plotted. In L929 cells exposed to EGCG, the expression levels of PPARγ, C/EBPα, aP2, and Pref-1 were significantly reduced to approximately 0.1–0.3-fold compared to the control (unexposed L929 cells) (p ≤ 0.05) (Fig. 6A). Conversely, in MA-3T3-L1 cells, PPARγ expression was significantly decreased (p ≤ 0.05), while C/EBPα, Pref-1, and aP2 expression levels showed no significant changes compared to the control (unexposed 3T3-L1 cells) (Fig. 6B).
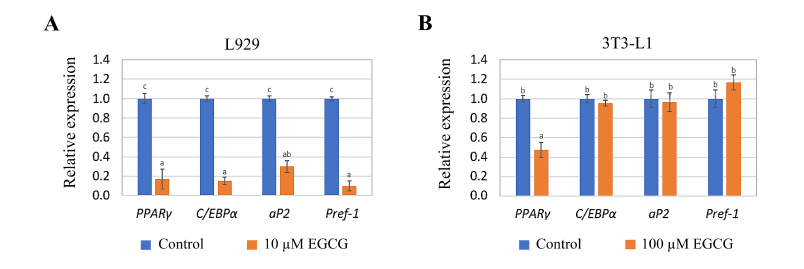 | Figure 6. Relative expression of adipogenesis-related genes in (A) L929 cells exposed to 10 µM EGCG for 3 days and (B) 3T3-L1 cells exposed to 100 µM EGCG for 14 days. The letters (a, b and c) at the top of the bars indicate significant differences in the relative expression levels among the genes within each cell type (p ≤ 0.05). [Click here to view] |
DISCUSSION
According to the spontaneous adipocyte differentiation potential of L929, this cell line was selected as an alternative model for evaluating anti-adipogenesis compared to the conventional 3T3-L1 cells in this study. EGCG was administered as a gold standard substance to investigate cellular sensitivity, anti-adipogenesis potential, and alterations in adipogenic-related gene expression. After 72 hours of the sensitivity test, L929 cells exhibited greater sensitivity to EGCG compared to 3T3-L1 cells, as evidenced by the dose-dependent decrease in L929 viability (p ≤ 0.05) (Fig. 2). Consistent with previous reports, exposure to 300 µg/ml (65.45 µM) EGCG for 6 hours reduced L929 viability [20], while high concentrations of EGCG under defined exposure times decreased the survival of 3T3-L1 cells [21]. It is conceivable that EGCG may exert varying effects on different cell types. Previous studies have demonstrated reduced viability and increased apoptosis in adipocytes exposed to EGCG [22-24], aligning with the sensitivity of L929 cells, originally derived from subcutaneous adipose tissue of a male C3H/An mouse [25].
Previous studies have shown that EGCG inhibits adipogenesis in 3T3-L1 cells [22,26,27]. EGCG dose-dependently inhibited lipid accumulation in maturing preadipocytes. The inhibitory effects of EGCG are mediated by PI3K-AKT signaling to down-regulate PPARγ and FAS expression levels at 100 µM [26]. A-dependent CCNA2 and CDK2 expressions mediated by FTO and YTHDF2 also contributed to EGCG-induced adipogenesis inhibition at 200 µM [27].
In contrast, 3T3-L1 cells, derived from a small cell cluster of mouse fibroblasts [28], were able to survive exposure to 680 µg/ml or approximately 1,483.51 µM EGCG for 2 days [21]. However, high concentrations of EGCG can alter cellular metabolism, leading to proliferation inhibition, apoptosis, and clastogenic induction. Additionally, EGCG’s oxidative reaction with molecules in the culture medium generates toxic H2O2 and other oxidation products, causing mitochondrial dysfunction, oxidative stress, and apoptosis [29-31].
The distinct origins of cells impart various properties, including adipocyte differentiation capacity, cell size, and lipid droplet volume. External factors also play a pivotal role in shaping cellular characteristics. As depicted in Figure 2, 3T3-L1 cells exhibited larger lipid droplets compared to those observed in L929 cells. The adipocyte induction medium for 3T3-L1 cells typically contains high concentrations of insulin, growth factors, and cytokines, facilitating the maturation of adipocytes over a 2-week period. These induction substances are recognized as crucial factors contributing to the enlargement of lipid droplets and are associated with insulin resistance in mature adipocytes [32-35]. Lipid droplet size and cellular composition also vary significantly between adipose tissue types. Brown-like adipose tissue (BeAT), often found in subcutaneous white fat depots, is characterized by numerous small lipid droplets and abundant mitochondria in the cytoplasm. This unique composition enhances its sensitivity to EGCG, which may influence its metabolic activity and lipid dynamics. Conversely, white adipose tissue (WAT) accumulates larger lipid droplets, contains fewer mitochondria, and exhibits lower metabolic activity, resulting in greater resistance to higher concentrations of EGCG [15]. These distinctions between BeAT and WAT have been shown to affect metabolic rate, adipogenesis, and adipolysis. The interplay between lipid droplet size, mitochondrial content, and adipose tissue type has been extensively documented as critical factors in these processes [36-38].
The effects of EGCG on adipogenesis inhibition in both cell types were corroborated by the heterogeneous expression levels of specific genes. Two distinct groups of genes were assessed: (1) adipocyte differentiation genes, including PPARγ, C/EBPα, and Pref-1; and (2) lipid metabolism genes, such as aP2. During adipocyte differentiation, PPARγ and C/EBPα are induced and upregulated, subsequently targeting fatty acid synthase and aP2 activation, leading to cytoplasmic lipid accumulation. Pref-1 serves as a regulatory molecule for both PPARγ and C/EBPα; its down-regulation is essential for the expression of C/EBPα and PPARγ, thereby facilitating adipogenesis [39]. Pref-1, also known as DLK1, is a key regulator in adipogenesis. Understanding its function is crucial for contextualizing studies on adipocyte differentiation. It inhibits the maturation of preadipocytes into adipocytes, preventing the accumulation of lipid droplets and the expression of adipogenic markers [40-42]. Pref-1 functions by binding to specific receptors on the cell surface and activating signaling pathways that inhibit the expression of key adipogenic transcription factors and this inhibition blocks the terminal differentiation of preadipocytes. The expression of Pref-1 decreases as preadipocytes differentiate into mature adipocytes [40-42]. This downregulation is necessary for the differentiation process to proceed. High levels of Pref-1 are associated with maintaining cells in an undifferentiated state. Pref-1’s role in regulating adipogenesis has implications for metabolic disorders, such as obesity and diabetes. Abnormal expression of Pref-1 can lead to alterations in fat tissue development and function. Understanding Pref-1’s function can also provide insights into potential therapeutic targets for controlling adipogenesis and related metabolic conditions [40-42].
In L929 cells, 10 µM EGCG was selected for this experiment due to a lack of reduction in cell viability with a reduction in lipid droplets observed at this concentration (Fig. 2 and 5). Following EGCG exposure, a significant decrease in the expression levels of all assessed genes was observed (Fig. 6A), suggesting that EGCG influenced gene expression. Conversely, in 3T3-L1 cells, the expression of PPARγ, C/EBPα, and aP2 decreased, while Pref-1 was unchanged (Fig. 6B). This resulted in the interruption of adipogenesis at the pre-adipocyte stage, characterized by a high expression level of Pref-1 in 3T3-L1 cells [9]. The observed discrepancies in the effects of EGCG on gene expression and lipid content between L929 cells and 3T3-L1 cells can be attributed to several factors, including differences in cell type, cellular context, and concentration response dynamics. L929 and 3T3-L1 cells originate from different tissues and have distinct physiological roles. L929 cells are fibroblast-like cells derived from mouse connective tissue, often used in cytotoxicity studies and general cell biology research. 3T3-L1 cells are preadipocyte cells derived from mouse embryo fibroblasts, widely used as a model for studying adipogenesis (fat cell formation). These intrinsic differences can lead to varied responses to the same compound, such as EGCG, due to differences in receptor expression, signal transduction pathways, and metabolic activities. The sensitivity of cells to EGCG can differ based on gene expression as L929 cells might express higher levels of receptors or proteins that are sensitive to EGCG, leading to a more significant downregulation of gene expression at lower concentrations (10–40 μM). 3T3-L1 cells, being adipogenic, have pathways more directly involved in lipid metabolism. Therefore, higher concentrations of EGCG (>200 μM) might be needed to observe a notable effect on lipid content. The effects of EGCG are often dose-dependent, and different concentrations can trigger distinct cellular responses. In L929 cells, a lower concentration might be sufficient to downregulate specific genes without appreciably affecting lipid metabolism, while in 3T3-L1 cells, higher concentrations may be necessary to influence both gene expression and lipid content significantly. EGCG might affect various cellular mechanisms in different cell types in L929 cells, it could be more effective in modulating gene expression pathways, such as those involved in cell proliferation, differentiation, or stress response, without impacting lipid content. In 3T3-L1 cells, EGCG might act more prominently on lipid metabolic pathways, leading to a reduction in lipid accumulation. Differences in experimental conditions, such as culture media, serum concentration, and incubation times, can also influence the outcomes. These conditions might affect the availability, stability, and uptake of EGCG in different cell types, thereby leading to varied results. The importance of considering cell-specific factors and dose-response relationships when interpreting the effects of compounds like EGCG are important. Further investigation, including detailed mechanistic studies, could help clarify the underlying reasons for these observed differences.
Numerous studies have demonstrated that EGCG regulates a plethora of genes involved in adipogenesis, lipolysis, beta-oxidation, and thermogenesis [43-45]. In male C57BL/6J mice fed with 0.2% or 0.5% EGCG (w/w) for 8 weeks, the mRNA levels of adipogenic genes, such as PPARγ, C/EBPα, aP2, LPL, and FAS, were significantly decreased [46]. Treatment with 5 µM EGCG for 3 and 9 days in 3T3-L1 cells also reduced lipid accumulation by approximately 84.1% compared to the control [47]. Green tea containing EGCG mediated adipocyte differentiation by suppressing the expression of Pref-1, PPAR-γ, C/EBPα, and C/EBP-β in perirenal fat [48]. The observed gene expression patterns in both L929 and 3T3-L1 cells, influenced by EGCG, indicate the interference with adipocyte differentiation and subsequent lipid accumulation, highlighting a consistent response to EGCG across both cell types at the gene expression level.
In the realm of adipogenesis research, a challenge lies in selecting techniques that swiftly and accurately screen fat accumulation in cells. While a myriad of methodologies exists, the key is finding approaches that balance simplicity with robustness in quantifying adipogenic outcomes. At times, substances have been observed to induce or inhibit the formation of fat droplets, yet there remains a lack of a rapid tool to effectively quantify the resulting fat accumulation numerically. Image analysis software such as ImageJ provides a promising solution by leveraging sophisticated image processing algorithms to enable precise quantification of cellular parameters [49]. This allows for objective and reproducible assessment of adipogenic responses. Additionally, our research introduces the utilization of L929 cells in conjunction with image-based analysis, offering a time-efficient and cost-effective approach with reasonable validity. By utilizing cell lines that bypass induction protocols and boast accelerated differentiation timelines, we streamline the screening process. Validation results confirm the feasibility and effectiveness of this approach, highlighting its potential as a valuable screening tool. Nonetheless, it is crucial to acknowledge potential limitations inherent to this methodology, such as imaging artifacts or background noise. These challenges can be mitigated by standardizing imaging conditions and optimizing parameters to minimize technical issues [50]. Additionally, the study relies solely on L929 cells, which, while efficient for adipocyte differentiation, may not fully replicate the adipogenesis process observed in other cell types. Validation of findings in additional models, such as primary pre-adipocytes or other cell lines, is necessary to ensure broader applicability.
Another limitation is the assessment of EGCG’s effects primarily in non-induced adipocytes, which may not fully represent the complex differentiation process occurring in pre-adipocytes. Investigating EGCG’s impact during the early stages of adipogenesis could provide a more comprehensive understanding of its mechanisms. Furthermore, the study lacks an in vivo component, which is essential for evaluating the systemic effects of EGCG, including its bioavailability, metabolism, and long-term impact on metabolic health. Future studies should employ more complex in vivo models to confirm these findings and explore the sustained effects of EGCG treatment.
While the image-based methodology offers efficiency, it may overlook detailed biochemical insights into lipid composition and turnover. Combining this approach with traditional biochemical assays could provide a more holistic view of lipid metabolism and adipogenesis. Moreover, it is important to note that this study presents a preliminary model intended as a practical screening tool. For more precise experimental outcomes, complementary investigations utilizing additional methodologies are essential.
CONCLUSION
In summary, our study demonstrated that the combined use of L929 cells and image-based assays offers a rapid and reliable approach for experiments related to adipogenic responses. This method eliminates the need for an adipogenic induction process and does not require a biochemical evaluation of lipid accumulation. The non-adipogenic induction process for L929 cells significantly shortens the processing time compared to the conventional 3T3-L1 method, which requires up to 14 days of induction. In contrast, L929 cells differentiated into mature adipocytes within just 5 days. Although the sensitivity to EGCG differed between L929 and 3T3-L1 cells due to their distinct cellular characteristics, the overall response to EGCG in terms of anti-adipogenesis and gene expression was consistent across both cell types. This study underscores the efficacy of L929 cells as a viable alternative model for adipogenesis research, streamlining the screening process while maintaining robust and reproducible results.
ACKNOWLEDGMENTS
The authors would like to express their deepest gratitude to Assistant Professor Dr. Weerah Wongkham for his invaluable guidance and support throughout this research project.
AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FINANCIAL SUPPORT
The authors gratefully acknowledge the research funds from the Faculty of Science and Technology, Thammasat University (No. 32/2560) to Kewalin Inthanon, and the Bualuang ASEAN Chair Professorship to Neal M. Davies.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All the data are available with the authors and shall be provided upon request.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Mejia-Meza EI, Yanez JA, Remsberg CM, Takemoto JK, Davies NM, Rasco B, et al. Effect of dehydration on raspberries: polyphenol and anthocyanin retention, antioxidant capacity, and antiadipogenic activity. J Food Sci. 2010;75(1):H5–H12.
2. Takemoto JK, Remsberg CM, Davies NM. Pharmacologic activities of 3’-hydroxypterostilbene: cytotoxic, anti-oxidant, anti-adipogenic, anti-inflammatory, histone deacetylase and sirtuin 1 inhibitory activity. J Pharm Sci. 2015;18(4):713–27.
3. Bahmad HF, Daouk R, Azar J, Sapudom J, Teo JC, Abou-Kheir W, et al. Modeling adipogenesis: current and future perspective. Cells. 2020;9(10):2326.
4. Maier-Hein L, Eisenmann M, Reinke A, Onogur S, Stankovic M, Scholz P, et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat Commun. 2018;9(1):5217.
5. Mori E, Fujikura J, Noguchi M, Nakao K, Matsubara M, Sone M, et al. Impaired adipogenic capacity in induced pluripotent stem cells from lipodystrophic patients with BSCL2 mutations. Metabolism. 2016;65(4):543–56.
6. Sririwichitchai R, Saiai A, Inthanon K, Chomdej S, Wongkham W, Roongruangwongse W. Anti-adipogenesis activities of Zingiber cassumunar Roxb. rhizome extracts on L929 cells evaluated by image-based analysis. Vet Integr Sci. 2018;16(2):35–51.
7. Paul T, Apte KG, Parab PB, Das B. Role of Adiantum philippense L. on glucose uptake in isolated pancreatic cells and inhibition of adipocyte differentiation in 3T3-L1 cell line. Pharmacogn Mag. 2017;13(Suppl 2):S334.
8. Theerakittayakorn K, Bunprasert T. Differentiation capacity of mouse L929 fibroblastic cell line compare with human dermal fibroblast. Int J Med Sci. 2011;5(2):51–4.
9. Jakab J, Miški? B, Mikši? Š, Jurani? B, ?osi? V, Schwarz D, et al. Adipogenesis as a potential anti-obesity target: a review of pharmacological treatment and natural products. Diabetes Metab Syndr Obes. 2021 Jan 8;14:67–83.
10. Peng H, Lin X, Wang Y, Chen J, Zhao Q, Chen S, et al. Epigallocatechin gallate suppresses mitotic clonal expansion and adipogenic differentiation of preadipocytes through impeding JAK2/STAT3-mediated transcriptional cascades. Phytomedicine. 2024;129:155563.
11. Zhang Z, Jia Y, Zhang C, Zhang Z, Jin F, Pan D, et al. Efficacy of epigallocatechin gallate (EGCG) and its underlying mechanism in preventing bisphenol-A-induced metabolic disorders in mice. J Hazard Mater. 2024;469:134098.
12. Jeney F, Bazsó-Dombi E, Oravecz K, Szabó J, Nagy IZ. Cytochemical studies on the fibroblast-preadipocyte relationships in cultured fibroblast cell lines. Acta Histochem. 2000;102(4):381–9.
13. Van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Biol. 2011;731:237–45.
14. Sakuma M. Probit analysis of preference data. Appl Entomol Zoolog. 1998;33(3):339–47.
15. Wong-a-nan N, Inthanon K, Saiai A, Inta A, Nimlamool W, Chomdej S, et al. Lipogenesis inhibition and adipogenesis regulation via PPARγ pathway in 3T3-L1 cells by Zingiber cassumunar Roxb. rhizome extracts. EJBAS. 2018;5(4):289–97.
16. Kinkel AD, Fernyhough ME, Helterline DL, Vierck JL, Oberg KS, Vance TJ, et al. Oil red-O stains non-adipogenic cells: a precautionary note. Cytotechnology. 2004;46:49–56.
17. Li Y, Sair AT, Zhao W, Li T, Liu RH. Ferulic acid mediates metabolic syndrome via the regulation of hepatic glucose and lipid metabolisms and the insulin/IGF-1 receptor/Pi3K/AKT pathway in palmitate-treated HepG2 cells. J Agric Food Chem. 2022;70(46):14706–17.
18. Broeke J, Pérez J MM, Pascau J. Image processing with Image J. 2nd ed. Birmingham, UK: Packt Publishing Ltd; 2015.
19. Livak K J, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8.
20. Matsumura K, Kim JY, Tsutsumi S, Hyon SH. Hibernation, reversible cell growth inhibition by epigallocatechin-3-O-gallate. J Biotech. 2007;127(4):758–64.
21. Lao W, Tan Y, Jin X, Xiao L, Kim JJ, Qu X. Comparison of cytotoxicity and the anti-adipogenic effect of green tea polyphenols with epigallocatechin-3-gallate in 3T3-L1 preadipocytes. Am J Chin Med. 2015;43(6):1177–90.
22. Lin J, Della-Fera MA, Baile CA. Green tea polyphenol epigallocatechin gallate inhibits adipogenesis and induces apoptosis in 3T3-L1 adipocytes. Obes Res. 2005;13(6):982–90.
23. Bécsi B, Kónya Z, Boratkó A, Kovács K, Erd?di F. Epigallocatechine-3-gallate inhibits the adipogenesis of human mesenchymal stem cells via the regulation of protein phosphatase-2A and myosin phosphatase. Cells. 2022;11(10):1704.
24. Chen CP, Su TC, Yang MJ, Chen WT, Siao AC, Huang LR. et al. Green tea epigallocatechin gallate suppresses 3T3-L1 cell growth via microRNA-143/MAPK7 pathways. EBM. 2022;247(18):1670–9.
25. Rawat SG, Tiwari RK, Sonker P, Maurya RP, Vishvakarma NK, Kumar A. EGCG as anti-obesity and anticancer agent. Book: Obesity and Cancer. Berlin, Germany: Springer Nature; 2021. pp. 209–33.
26. Rodriguez T, Rengifo E, Gavilondo J, Tormo B, Fernández A. Morphologic and cytochemical study of L929 cell variants with different metastasizing ability in C3HA/Hab mice. Neoplasma. 1984;31(3):271–9.
27. Wu M, Liu D, Zeng R, Xian T, Lu Y, Zeng G, et al. Epigallocatechin-3-gallate inhibits adipogenesis through down-regulation of PPARγ and FAS expression mediated by PI3K-AKT signaling in 3T3-L1 cells. Eur J Pharmacol. 2017;795:134–42.
28. Wu R, Yao Y, Jiang Q, Cai M, Liu Q, Wang Y, et al. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int J Obes (Lond). 2018;42(7):1378–88.
29. Green H. U.S. Patent No. 4,003,789. Washington, DC: U.S. Patent and Trademark Office; 1977, p. 697.
30. Chen B, Zhang W, Lin C, Zhang L. A comprehensive review on beneficial effects of catechins on secondary mitochondrial diseases. Int J Mol Sci. 2022;23(19):11569.
31. Mokra D, Joskova M, Mokry J. Therapeutic effects of green tea polyphenol (?) -epigallocatechin-3-Gallate (EGCG) in relation to molecular pathways controlling inflammation, oxidative stress, and apoptosis. Int J Mol Sci. 2022;24(1):340.
32. Ouyang J, Zhu K, Liu Z, Huang J. Prooxidant effects of epigallocatechin-3-gallate in health benefits and potential adverse effect. Oxid Med Cell Longev. 2020;1:9723686.
33. Ahmed B, Sultana R, Greene MW. Adipose tissue and insulin resistance in obese. Biomed Pharmacother. 2021;137:111315.
34. Czech MP. Mechanisms of insulin resistance related to white, beige, and brown adipocytes. Mol Metab. 2020;34:27–42.
35. Henne WM, Reese ML, Goodman JM. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 2018;37(12):e98947.
36. Remsberg CM, Martinez SE, Akinwumi BC, Anderson HD, Takemoto JK, Sayre CL, et al. Preclinical pharmacokinetics and pharmacodynamics and content analysis of gnetol in foodstuffs. Phytother Res. 2015;29(8):1168–79.
37. Brownstein AJ, Veliova M, Acin-Perez R, Villalobos F, Petcherski A, Tombolato A, et al. Mitochondria isolated from lipid droplets of white adipose tissue reveal functional differences based on lipid droplet size. Life Sci Alliance. 2024;7(2):e202301934.
38. Bosch M, Parton RG, Pol A. Lipid droplets, bioenergetic fluxes, and metabolic flexibility. Semin Cell Biol. 2020;108:33–46.
39. Veliova M, Petcherski A, Liesa M, Shirihai OS. The biology of lipid droplet-bound mitochondria. Semin Cell Dev Biol. 2020;108:55–64.
40. Sun C, Mao S, Chen S, Zhang W, Liu C. PPARs-orchestrated metabolic homeostasis in the adipose tissue. Int J Mol Sci. 2021;22(16):8974. doi: https://doi.org/10.3390/ijms22168974
41. Sul HS, Smas C, Mei B, Zhou L. Function of pref-1 as an inhibitor of adipocyte differentiation. Int J Obes Relat Metab Disord. 2000;24 Suppl 4:S15–9.
42. Lee MS, Kim CT, Kim Y. Green tea (–)-epigallocatechin-3-gallate reduces body weight with regulation of multiple genes expression in adipose tissue of diet-induced obese mice. Ann Nutr Metab. 2009;54(2):151–7.
43. Wang Y, Kim KA, Kim JH, Sul HS. Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J Nutr. 2006;136(12):2953–6.
44. Gu Q, Xia L, Du Q, Shao Y, He J, Wu P, et al. The therapeutic role and potential mechanism of EGCG in obesity-related precocious puberty as determined by integrated metabolomics and network pharmacology. Front Endocrinol. 2023;14:1159657.
45. Tahri-Joutey M, Andreoletti P, Surapureddi S, Nasser B, Cherkaoui-Malki M, Latruffe N. Mechanisms mediating the regulation of peroxisomal fatty acid beta-oxidation by PPARα. Int J Mol Sci. 2021;22(16):8969.
46. Faria A, Pereira-Wilson C, Negrão R. The relevance of polyphenols in obesity therapy. In Monteiro R (Ed.), Martins MJ (Co-Ed.). Recent advances in obesity: understanding obesity - from its origins to impact on life. Bentham Science Publishers; 2020. pp. 271–307.
47. Lee K, Villena JA, Moon YS, Kim KH, Lee S, Kang C, et al. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J Clin Invest. 2003;111(4):453–61.
48. Furuyashiki T, Nagayasu H, Aoki Y, Bessho H, Hashimoto T, Kanazawa K, et al. Tea catechin suppresses adipocyte differentiation accompanied by down-regulation of PPARγ2 and C/EBPα in 3T3-L1 cells. Biosci Biotechnol Biochem. 2004;68(11):2353–9.
49. Chen N, Bezzina R, Hinch E, Lewandowski PA, Cameron-Smith D, Mathai ML, et al. Green tea, black tea, and epigallocatechin modify body composition, improve glucose tolerance, and differentially alter metabolic gene expression in rats fed a high-fat diet. Nutr Res. 2009;29(11):784–93.
50. Dobson ET, Cimini B, Klemm AH, Wählby C, Carpenter AE, Eliceiri KW. ImageJ and CellProfiler: complements in open-source bioimage analysis. Curr Protoc. 2021;1(5):e89.
51. Caicedo JC, Cooper S, Heigwer F, Warchal S, Qiu P, Molnar C, et al. Data-analysis strategies for image-based cell profiling. Nat Methods. 2017;14(9):849–63.