INTRODUCTION
Parinari curatellifolia (PC) Planch. Ex Benth. (Family: Chrysobalanaceae), commonly known as Mobola plum, is an evergreen tropical tree plant that is native to most parts of sub-Saharan Africa including Nigeria, Kenya, and Zimbabwe [1]. Over time, it has been noted to be a multipurpose plant with numerous nutritional, medicinal, and industrial benefits. It has been found significant in human nutrition as the tree produces delicious plum-size fruits with yellow flesh which is edible both in raw and dried form [2]. It has been noted to have a pleasant, sweet taste and it is considered by many as one of the most desirable African wild fruits [3]. In addition, the seed kernel possesses high oil content, hence its edible oil is also used for cooking [2].
Traditionally, many parts of P. curatellifolia have been used to treat various diseases across Africa [1,2]. Records have shown that the leaf, stem, and bark are used in Nigeria as traditional remedies for cough, diabetes, nervous disorders, piles, and skin infections [4,5]. For instance, the plant has been documented in South African ethnomedicine as an effective treatment option for skin disorders [4] and it was particularly identified as one of the plants with the highest “use rate” among the Mapulana people of the country [5]. Thus, different scientific studies have shown that the plant and its constituents possess significant biological activities including analgesic [6], antioxidant [7,8], anticancer [9], anti-microbial [8], cardioprotective [10], and hepatoprotective [1,11], to mention a few.
In particular, the hepatoprotective effects of P. curatellifolia are remarkable judging from both its acclaimed folkloric uses and supporting scientific evidence. Atawodi et al. [11] noted that P. curatellifolia was widely used in the traditional management of liver-related illnesses among the Hausa tribe of Northern Nigeria. Scientific research has further demonstrated that extracts from the plant’s roots significantly protected the liver against carbon tetrachloride-induced oxidative stress and liver damage [12]. Subsequently, the methanolic extracts of P. curatellifolia roots were also shown to display hepatoprotective effects on both acute and chronic liver damage induced in rats by carbon tetrachloride [11].
Previous studies in our laboratory, which were focused on the seeds, had also revealed the potential of P. curatellifolia in the treatment of liver damage. The seeds were selected for study based on ethnomedical records and our findings that P. curatellifolia seed extracts had remarkable anti-oxidative potential [1]. In addition, the seeds were selected for study because silymarin, which is the standard therapeutic for hepatic disorders, is a lipophilic extract from the seeds of milk thistle [13]. Previous results also showed that the lipophilic flavonoid-rich extract (FRE) of the Parinari seeds was principally responsible for its inherent anti-hepatoxic activities [1], which are in consonance with studies that showed that a flavonoid, silibinin is the major component of silymarin mixture responsible for its hepatoprotective effects [14]. Interestingly, P. curatellifolia seeds have also been documented to contain a variety of other phenolics compounds and these Parinari flavonoids have been shown to be therapeutically effective against several debilitating conditions such as atherogenicity and dyslipidaemia [15].
Generally, the study of flavonoids in plants has stimulated considerable interest due to their numerous health benefits, with novel flavonoids with diverse biological activities continually being sourced from both food and medicinal plants [16]. Scientific evidence has also shown that the dietary intake of flavonoids has varying ameliorative effects against diseases related to free radical generation such as cardiovascular diseases, diabetes, hypertension, as well as liver, kidney, and brain damage [17]. A very remarkable example of such important flavonoids is the earlier mentioned flavonoid-rich silymarin (the extract of milk thistle) and its major active compound, silibinin, which have been identified as standard therapeutics in different liver disorders including chronic liver diseases cirrhosis and hepatocellular carcinoma [18]. Hence, the a need to investigate other flavonoids from various plants with documented hepatoprotective activity such as P. curatellifolia, and establish their activities.
Thus, this study aimed to highlight the flavonoid components of the hepatoprotective P. curatellifolia seed extracts and to evaluate these compounds using in-silico approaches to facilitate the discovery of anti-hepatotoxic lead compounds. In silico approaches are widely used in drug discovery, especially at the initial stages of drug discovery and development, to evaluate the pharmacokinetics, biological, and toxic properties of compounds, mainly because of their speed of analysis and cost efficiency [19]. Hence, the identified flavonoids from P. curatellifolia were subjected to bioactivity prediction, molecular properties prediction, toxicity prediction, and drug-likeness using various web-based tools. This research also elucidates one of the probable interactive mechanisms at work between selected P. curatellifolia flavonoids and CYP1A2 (Cytochrome P450 family 1 subfamily A member 2), using silibinin as the standard. To the best of our knowledge, this is the first study describing the correlation between the hepatoprotective activities of P. curatellifolia and its flavonoid constituents using a computational approach.
MATERIALS AND METHODS
Plant collection
PC seeds were purchased at a local market in Akure, Nigeria. Subsequently, the botanical identification and authentication of the plant materials were carried out and the plant authentication number (PC005) was deposited in the Department of Biochemistry, Federal University of Technology Akure, Nigeria.
Preparation of FREs
The PC seeds were air-dried to constant weight, pulverized using an industrial blender, and stored in airtight containers before usage. Crude extraction of the seeds was carried out with methanol (80%) using a Soxhlet apparatus and the solvent was removed using a rotary evaporator. Subsequently, the crude extract (6 g) was dissolved in 40 ml of 1% H2SO4 and hydrolyzed by heating in a water bath for 30 minutes at 70°C. The resulting mixture was cooled on ice for 15 minutes to facilitate the precipitation of flavonoid aglycones. The solution was filtered and the solid (flavonoid aglycone mixture) was dissolved in 100 ml of ethanol (95%) at 50°C. The resulting solution was filtered into a 200 ml volumetric flask and made up to the mark with 95% ethanol. The filtrate was finally concentrated to dryness using a rotary evaporator [1].
Phytochemical screening
Phytochemical screenings of both the crude and FREs were done to determine the constituents, using standard methods [20,21]. The phytochemical analyses included the identification of alkaloids, anthraquinones, cardenolides, flavonoids, phlobatannins, saponins, terpenoids, and tannins.
High performance liquid chromatography (HPLC) analysis of the FRE
The HPLC fingerprinting of the FRE of P. curatellifolia seeds was performed through HPLC-DAD (Agilent Technologies, USA) under gradient conditions using a Phenomenex C-18 column following the method of Öztürk, Tunçel, and Poto?lu-Erkara [22]. The mobile phase used was solvent A = water/acetic acid (99:1) and solvent B = acetonitrile. The Parinari flavonoid extract was dissolved in ethanol (10 mg/ml) and analyzed for the presence of 3-methylquercetin (3MQ), apigenin, baicalein, chrysin, genistein, isorhamnetin, kaempferol, myricetin, pinocembrin, and quercetin. Commercial standards served as control (Sigma, USA).
Physicochemical properties and bioactivity score
The physicochemical properties of the flavonoid compounds and silibinin (standard), were predicted using the Molinspiration physicochemical properties calculator (www.molinspiration.com). The properties evaluated include the number of heavy atoms (nAtoms), number of hydrogen donors (nHOH), number of hydrogen acceptors (nON), the molecular weight (M.wt), the partition coefficient (log P), the topological polar surface area (TPSA), and the number of violations (nviol) [23]. In addition, the bioactivity scores of the flavonoids were computed by estimating the activity scores for ion channel modulator, kinase inhibitor, G protein-coupled receptors (GPCRs) ligand, and nuclear receptor ligand using the Molinspiration drug-likeness platform [24].
Prediction of activity spectra for substances (PASS) bioactivity prediction
Predictions of the biological potentials of the identified P. curatellifolia flavonoids were performed using the PASSs program which is available at www.pharmaexpert.ru/passonline/as earlier reported [25].
Toxicity risk prediction
Computational simulation studies were carried out to estimate the potential toxicity risks of the flavonoids using three online computer programs: admetSAR platform (www.lmmd.ecust.edu.cn/admetsar2), PreADMET (https://preadmet.bmdrc.kr/), and the OSIRIS Property Explorer (www.organic-chemistry.org/prog/peo/). The evaluated risks included cardiotoxicity, carcinogenicity, eye irritability, mutagenicity, and reproductive side effects. The data were analyzed and expressed as low-risk, medium-risk, high-risk, and non-detected risk levels [25].
Preparation of enzyme and ligands
The X-ray crystal structure of CYP1A2 (PDB code: 2HI4) was obtained from the RSCB Protein Data Bank and prepared by removing nonstandard residues, protein connectivity, and water molecules using the ChimeraX software [26]. The structures of the flavonoids and the standard (silibinin) were retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/) and their 3-D structures were optimized using Avogadro software by the inclusion of Gasteiger charges and the non-polar hydrogen. Thereafter, the resulting optimized flavonoids and CYP1A2 were subjected to molecular docking.
Molecular docking
Docking was executed using the Autodock Vina Plugin on Chimera. Briefly, the optimized flavonoids were docked into the binding pockets of the target, Cytochrome P450 1A1 CYP1A2 (grid box spacing of 1 Å and size of 24 × 30 × 29 in x, y, and z directions, respectively). Judging by the docking scores, the docked complexes with the best pose were subsequently subjected to molecular dynamics simulations (MDSs).
MDS
The MDS of the selected compounds from section 2.8 was carried out as previously detailed, using the graphics processing unit version with the AMBER 18 package. ANTECHAMBER was employed in generating atomic partial charges for the flavonoids using the general amber force field and the restrained electrostatic potential procedures [27]. The Leap module of AMBER 18 allowed for the addition of H+ atoms as well as Cl- and Na+ counter ions for systems neutralization. The amino acid residues were numbered 1–495 for CYP1A2. The system was then taken through standard minimization (1,000 steps) and heating (0 to 300 K) through a 60 ns run [28]. Using PTRAJ, the resulting coordinates were saved while the trajectories were analyzed per ps before root mean square deviation (RMSD), root means square fluctuation (RMSF), and the radius of gyration (RoG) analyses by the C++ Processing of TRAJectories module. The binding affinity, the solvent accessible surface area (SASA), and the free binding energy (ΔG) for the individual specie (ligand, receptor, and complex) were computed and averaged over 100,000 snapshots extracted from the 60 ns trajectory [29].
Statistical analysis
The toxicity risk prediction data was presented as a heatmap constructed using the open-source web tool ClustVis (Metsalu and Vilo, 2015). The ΔGbind are presented as mean ± standard deviation of triplicate determinations, furthermore, the OriginPro V9.10 software was used for the MDS raw data plots. All differences were considered statistically significant if p < 0.05.
RESULTS
Parinari curatellifolia phytochemical constituents
The percentage yield of the FRE was found to be ~0.8% of the initial P. curatellifolia biomass. The phytochemical screening revealed that the FRE contained only flavonoids, while the methanolic crude extract contained alkaloids, cardenolides, flavonoids, phlobatannins, saponins, steroids, tannins, and terpenes (Supplementary Table 1). Furthermore, HPLC fingerprinting of P. curatellifolia FRE confirmed the presence of flavonoid compounds in different proportions. Specifically, the presence of 3-MQ, apigenin, baicalein, chrysin, genistein, isorhamnetin, kaempferol, myricetin, pinocembrin, quercetin, and tricetin were identified (Table 1). However, it was observed that kaempferol (39.17 mg/g), tricetin (33.13 mg/g), quercetin (31.41 mg/g), chrysin (25.57 mg/g), 3-MQ (21.86 mg/g), and pinocembrin (20.87 mg/g) are the most abundant flavonoids in P. curatellifolia FRE.
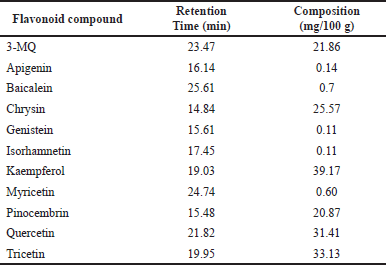 | Table 1. HPLC profile of PC flavonoids. [Click here to view] |
Molinspiration drug likeness
All the flavonoids identified from P. curatellifolia in this study fully comply with Lipinski’s rules; hence, they possess huge potential to serve as possible therapeutics or lead compounds (Table 2). The nine compounds in this study had LogP values ranging between 1.34 and 2.94, thus fulfilling Lipinski’s rule of ≤ 5; interestingly a few of the flavonoids compete more favorably than the standard, silibinin (LogP = 1.47) (Table 2). The molecular weights of all the compounds were also found to be 254.24–318.24 g/mol, which is below Lipinski’s 500 g/mol cut off, and also significantly lower than the standard hepatoprotective therapeutic, silibinin (M.wt = 482.44). Furthermore, it was found that the number of hydrogen bond acceptors (O and N atoms) and donors (NH and OH) for all the flavonoids were below five and ten, respectively. This observation agrees with Lipinski’s rule, and it must also be highlighted that the compounds performed better than silibinin in this regard. Results from this study showed that all the flavonoids in this study have acceptable TPSA values, all falling within the proposed values of 26–225, and were comparable to that of the standard.
Molinspiration bioactivity score
It is considered that the probability of a compound exhibiting any of the bioactivities predicted by the Molinspiration server becomes higher with increasing bioactivity score. Thus, a molecule with a bioactivity score greater than 0.00 is most likely to have significant biological activities; those with values between −0.50 and 0.00 are expected to exhibit moderate activities while those with <−0.50 are predicted to be inactive [30]. Similar to the standard drug silibinin, all the flavonoids evaluated in this study were predicted with a high probability of displaying their therapeutic ability mainly by acting as enzyme inhibitors and nuclear receptor ligands (Table 3). In addition, most of the flavonoids evaluated were predicted to also have the potential of being kinase inhibitors except apigenin and pinocembrin which had scores of −0.26 and −0.32, respectively. The negative scores recorded with the GCPR ligand, ion channel modulators, and protease inhibitor, however, suggest that the Parinari compounds do not utilize this mechanism of action for their respective activities.
PASS
The PASS analysis further revealed the specific biological activities and highlighted the other probable mechanisms responsible for the documented hepatoprotective activities of P. curatellifolia in mammals (Supplementary data: Table 2a-l). The specific biological activities directly related to protecting the liver against damage observed in most of the P. curatellifolia compounds include aldehyde oxidase inhibition, antioxidant activity, free radical scavenging ability, CYP1A induction, membrane integrity promotion, chemoprotection and peroxidase inhibition (Supplementary data: Table 2a-l). In addition, some other notable activities/mechanisms observed specifically in some flavonoids include histamine release inhibition in apigenin (Pa = 0.791), similarly, baicalein, genistein, and tricetin were predicted to possess anaphylatoxin receptor antagonism activities with Pa = 0.965, 0.801, and 0.945, respectively. Likewise, in this study, isorhamnetin was also predicted with high NOS-2 (nitric oxide synthase) expression inhibition (Pa = 0.895).
In silico toxicity risk prediction
The toxic risks assessment, viz., the cardiotoxicity, carcinogenicity, mutagenicity, eye irritability as well as the reproductive system toxicity (RST) of the flavonoids were predicted and represented in the heatmap (Fig. 1). While silibinin was used as the positive standard, carbon tetrachloride (CCl4), a known toxicant used frequently to induce toxicity in the liver and in many other organs, was used as the negative control [31]. All of the evaluated compounds were predicted to possess very little or no toxic potential when compared to CCl4. Particularly, all the flavonoid compounds displayed zero reproductive toxicity; however, quercetin was predicted to have a medium risk potential as a mutagen.
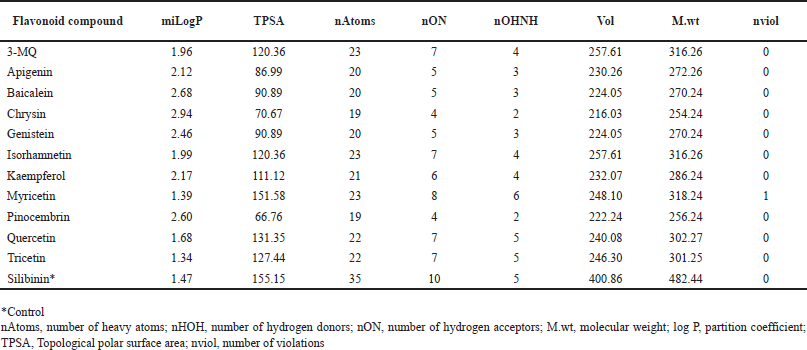 | Table 2. Drug likeness scores of P. curatellifolia flavonoids. [Click here to view] |
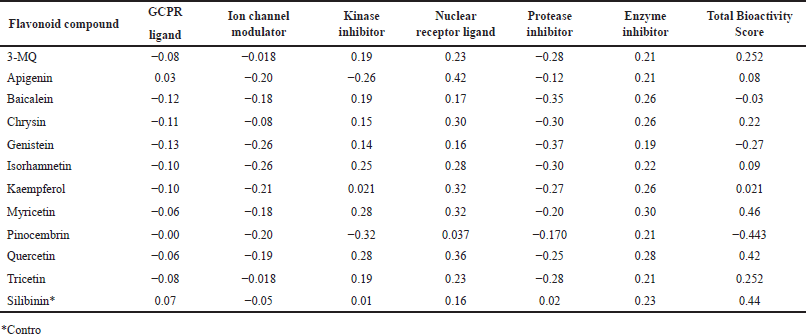 | Table 3. Bioactivity score of P. curatellifolia flavonoids. [Click here to view] |
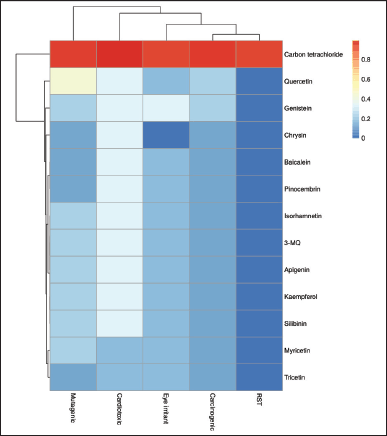 | Figure 1. Hierarchical clustering of the toxicity risk potential of P. curatellifolia flavonoids. Red colors indicate values greater than the substrate mean (increasing toxicity), while blue colors indicate values less than the mean (increasing safety). *RST = Reproductive system toxicity. [Click here to view] |
Molecular docking
Thus, the hepatoprotective potentials of the Parinari flavonoids to act as CYP1A1 inducers were evaluated through molecular docking. The results showed that the eleven compounds had binding energy scores ranging from −8.7 to −10.2 kcal/mol while the compound, silibinin had a lower binding energy of −10.7 kcal/mol (Table 4). However, chrysin and pinocembrin were found to compete most favorably with silibinin, as they both demonstrated the highest binding affinities (−10.2 kcal/mol) among all the evaluated flavonoids.
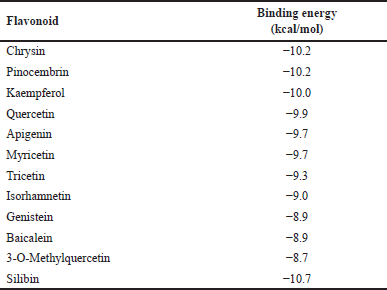 | Table 4. Energy scores of docking conformations of cytochrome P450 1A1 with P. curatellifolia flavonoids. [Click here to view] |
Molecular dynamic simulation refinement
The evaluated compounds demonstrated varying robust binding affinities (ΔGbind) between −35.85 and −32.00 kcal/mol; however, these binding affinities were lower when compared with the standard silibinin (ΔGbind = −58.52 kcal/mol) (Table 5). Furthermore, the computed binding energies were disintegrated to elucidate the nature of the predominant component interactions (Table 5). It was observed that similar to silibinin, the binding energies of all the P. curatellifolia flavonoids were dominated by ΔEvdW interactions, which is attributed to the van der Waals contribution. However, the ΔEvdW of the standard was −63.07 kcal/mol, which was significantly higher relative to those of the selected flavonoids between −38.23.54 and −35.93 kcal/mol. We also observed seven hotspot residues in the protein target, including Thr 91, Phe 92, Phe 193, Asn 279, Gly 283, Ala 284, and Thr 288, which exhibited effective interaction with all the tested flavonoids including the standard (Fig. 2a–e).
Furthermore, post-MD-based analyses were carried out to highlight the energetic and structural stabilities of the ligand-enzyme interactions. In this regard, the RMSD, RMSF, RoG, and SASA were evaluated. Assessment of the RMSD gives an indication of the structural changes in CYP1A1 during its interaction with the selected Parinari compounds. Results showed that the alpha carbon atoms for all the docked complexes achieved system equilibrium after 15 ns of the simulation (Fig. 3A). Furthermore, it was observed that the RMSD values steadily increased from 0 to 10 ns and reached equilibration after 15 ns while relatively few oscillations were observed subsequently throughout the simulation period (Fig. 3A). It was also observed that the free protein and the protein-flavonoid complexes displayed the same trend of fluctuations throughout the simulation. The average RMSD observed in the apo-enzyme and complex enzyme-flavonoid complexes were approximately 1.5 and 2.0 nm, respectively, which indicated slight structural deviation in the complex forms. However, there was no significant structural deviation between the apo structure and the complexes formed with chrysin, pinocembrin, and quercetin. Furthermore, besides kaempferol, all the evaluated P. curatellifolia flavonoids displayed lower RMSD when compared to silibinin, hence demonstrating higher consistency of energetic and structural measurements and consequently higher complex stability than the standard.
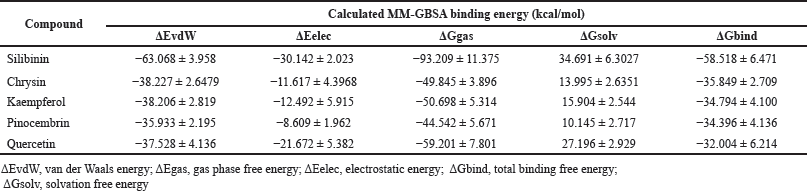 | Table 5. Calculated average MM-GBSA binding energies and corresponding energy components for P. curatellifolia flavonoids and silibinin in complex with CYP1A1. [Click here to view] |
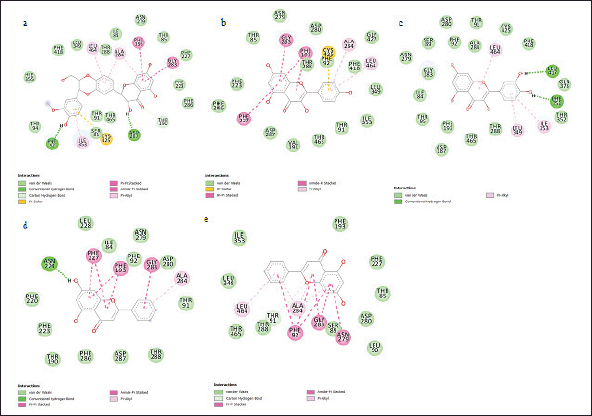 | Figure 2. Interaction plots of silybin (a) and P. curatellifolia flavonoids chrysin (b), kaempferol (c), pinocembrin (d), and quercetin (e) with CYP1A1 active site residues. [Click here to view] |
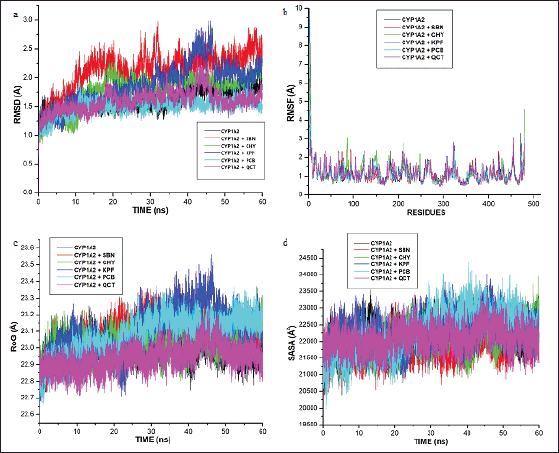 | Figure 3. Comparative alpha-carbon representations of the free cytochrome P450 1A1 (CYP1AI) and CYP1AI-P. curatellifolia flavonoid complexes during 60 ns simulation run illustrated as (a) root-mean-square deviation (RMSD), (b) root-mean-square fluctuation (RMSF), (c) RoG, and (d) SASA. CHY-Chrysin, KPF-Kaempferol, PCB-Pinocembrin, QCT-Quercetin, and SBN-Silibinin. [Click here to view] |
The RMSF data from this study show that the enzyme-flavonoid complexes formed were stable and that there were no significant fluctuational changes in the complexes relative to the free CYP1A1 enzyme (Fig. 3b). The mean RMSF values of the residues were found in the region of 0.8–2.5 Å for all the complexes while values of 0.8–2.9 Å were recorded for the apo structure, suggesting no significant changes in the evaluated systems.
In this study, the RoG values for the complexes and the unbounded enzyme demonstrate the same trend of compactness as they were found to be stable between 22.7–23.5 Å during the 60 ns simulation run, with only marginal differences (Fig. 3c). However, the complexes formed with quercetin and kaempferol were observed to display slight fluctuation between 42 and 48 ns (Fig. 3c). The average SASA values for the complexes were observed to be between 22,000 and 23,500 Å2 (Fig. 3d).
DISCUSSION
A significant quantity of flavonoids were recorded from the seeds of P. curatellifolia, although flavonoids are more concentrated in flowers and leaves, they are also found in significant quantities in seeds. For instance, the widely acclaimed silymarin is usually sourced from the fruits and seeds of milk thistle [32]. Previously, P. curatellifolia seeds crude extract have also been shown recently to contain the same classes of secondary metabolites identified in this study [15]; furthermore, our results are in line with a previous study that demonstrated kaempferol and quercetin as the prominent flavonoid compounds in P. curatellifolia FREs [15]. However, flavonoids such as 3-MQ, apigenin, baicalein, chrysin, genistein, isorhamnetin, myricetin, pinocembrin, and tricetin, are reported in P. curatellifolia, for the first time in this study. Lipinski’s drug-likeness rule of five states that for a compound to be considered as a possible drug candidate, it must possess hydrogen bond donor (nHOH) ≤ 5, hydrogen bond acceptor (nON) ≤ 10, (LogP) ≤ 5, molecular weight (M.wt) ≤ 500 Da with only one violation allowed [23]. Furthermore, compounds that do not comply with more than one of these rules lack the required activity and selectivity, making them less likely to be orally active in humans, and hence cannot be considered as potential drug candidates. It was observed that all the Parinari compounds in this study did not violate any of the rules and are more likely to serve as efficient therapeutics, especially in humans. For instance, P. curatellifolia flavonoids are predicted from the LogP values recorded in this study (Table 2) to possess significant permeability, and consequently, adequate lipid solubility to facilitate their interaction with the phospholipid bilayers and elicit bioactivity. LogP is a measure of the partition coefficient between n-octanol and water indicating a classical description of hydrophobicity and lipophilicity, thus, the solubility of a compound in water greatly facilitates activities related to drug development as well as the handling and formulation of the drug [33]. In the same vein, TPSA is a computed estimate of the surface area necessary to bind with most of the target receptors. It is considered as a notable factor to describe drug absorption, including bioavailability, intestinal absorption, permeability as well as blood-brain barrier penetration, thus, values between 26 and 225 have (Table 2), been noted to be necessary for a compound to exhibit satisfactory drug absorption [34]. Results from this study showed that all the flavonoids in this study have acceptable TPSA values, comparable to that of the standard, silibinin.
The Molinspiration bioactivity scores of our compounds of interest revealed their probable mechanism of therapeutic activity. Nuclear receptors have previously been identified among the most popular drug targets for human diseases including but not limited to cancer, cardiovascular diseases, diabetes, and especially liver damage [35]. Hence, as predicted in Table 3, a growing number of nuclear receptor ligands have been identified to control cellular homeostasis in the liver and other tissues, with their importance being demonstrated in cholestatic liver diseases [36] and non-alcoholic fatty liver disease (NAFLD) [37]. In the same vein, many compounds with enzyme-inhibiting activities, such as the SGLT-2 (Sodium-glucose cotransporter type-2) inhibitors have demonstrated remarkable efficacy in NAFLD by modulating and preventing pathways that lead to liver inflammation and apoptosis [38]. Based on the cumulative bioactivity scores (Table 3), it could be argued that many of the P. curatellifolia flavonoids compete favorably with silibinin; in fact, myricetin has a higher total score than the standard compound. In addition, results from this study demonstrate that the identified flavonoids probably display their therapeutic activities on liver damage via multiple mechanisms, including acting as GPCR ligands, nuclear receptor ligands, ion channel modulators, as well as inhibiting kinases, proteases, and other enzymes.
The PASS program computes the probability (Pa) of a compound to exhibit different biological activities, thus, Pa > 0.7 is considered as an indication that the evaluated compound will presumably display bioactivity in wet-lab experiments [39]. Many of these activities/ mechanisms highlighted in the Parinari flavonoids were also observed in silibinin, the standard drug, viz., free radical scavenging abilities, membrane integrity promotion, and antioxidant activity (Supplementary data: Table 2a-l). Recently, anaphylatoxin receptor antagonists have been demonstrated to protect against severe hepatic failure due to systemic inflammation and hepatic injury [40]. This is significant as inhibitors of NOS-2, such as NG-nitro-l-arginine methyl ester, have been proposed as anti-inflammatory agents, mainly because of inhibition of exacerbated NO formation in liver damage in malathion-intoxicated rats [41]. Hence, the promising probability values recorded in this study further indicate that P. curatellifolia compounds have a wide range of pharmacological activities and possible targets against specific receptors related to hepatoprotection.
While most flavonoids have been documented to be safe, the chemopreventive use of flavonoid therapy needs to be evaluated together with the toxicity. This is imperative as there have been some reports of toxic flavonoid-drug interactions, liver failure, contact dermatitis, hemolytic anemia, and reproductive health concerns associated with dietary flavonoid/phenolic consumption [42]. Compared to the negative control (CCl4), the Parinari flavonoids exhibited none of the toxicity assessed in this study except for quercetin which exhibited low potential of being a mutagen. It should be noted that the mutagenic potential of quercetin has been predicted in a previous study [43]. In all, it can be predicted that the identified flavonoids from P. curatellifolia possess very low risks and can be employed as therapeutics against liver damage or any other disease for that matter.
The activation of Cytochrome P450 1A1 (CYP1A1) has been directly linked to the detoxification of xenobiotics in the liver via metabolic activation, induction of phase I and II enzymes, as well as the reversal of the detrimental effects of lipid peroxidation: consequently, enhancing liver protection [44, 45]. CYP1A induction was also identified earlier in this study by the PASS bioactivity tool as a major metabolic route for the anti-hepatoxic potential of all the Parinari flavonoids (Supplementary data: Table 2a-l). The binding scores of the eleven flavonoids with the protein target, CYP1A, demonstrated the favorable interaction of the compounds with the active site region of the molecular target. Our previous had highlighted the hepatoprotective role of P. curatellifolia in acetaminophen-induced liver damage [1], which supports the findings that CYP1A1 is the probable cytochrome P450 isoform responsible for acetaminophen metabolism [46].
Further into the study, the ligands with the four lowest binding energies and consequently, the highest binding affinities for CYP1A1, viz., chrysin, kaempferol, pinocembrin, and quercetin, were selected for molecular dynamic simulation. This is necessary as the ligand-protein binding energies obtained through molecular docking scores are only preliminary and limited due to the lack of ligand-receptor flexibility, solvent effects, and dynamics [47]. Although the binding affinities (ΔGbind) of the selected compounds were lower than that of silibinin, they demonstrate high feasibility of either activating or inhibiting the molecular target. In this regard, they may be further optimized to perform even better than silibinin. As observed in this study, the dominance of van der Waals has been highlighted previously in many studies involving flavonoids and other protein targets [48]. Other significant interactions between the CYP1A1 binding site and the hepatoprotective compounds include pi-anion, pi-alkyl, and H-bond interactions. The significant binding of the Parinari compounds with the cytochrome was also established by the RMSD, RMF, RoG, and SASA plots (Fig. 3a–d). The RMSD observed in CYP1A1-flavonoid complexes were around 2.0 nm, thus indicating the remarkable stability of all the complexes formed. This observation is consistent with the reports of previous authors, who have established that lower RMSD values signify higher structural stability of an enzyme-ligand system [49,50]. The RMSF computes the magnitude of fluctuations along the principal axis, and it is useful in estimating the flexibility and movement of individual residues in the target protein during simulations [51]. The RMSF values in this study demonstrate that the binding of the hepatoprotective metabolites with the target protein did not affect the fluctuations of amino acid residues compared to free protein and the standard drug, silibinin, thus signifying limited movement during simulations and hence, structural stability. It has previously been shown that higher RMSF values indicate elevated movement of protein residues, while low values depict the restricted movement of the residues, implying robustness in the generated structures [52]. In addition, the flexibility of the activation loop was also evaluated by RMSF (Fig. 3b). The region possessing higher flexibility in the target protein is identified by comparing the RMSF of all the residues throughout the simulation trajectory and this gives an insight into the probable binding pockets and the active site [51]. As observed in the RMSF plot, it was found that the major peaks of fluctuation were almost evenly distributed along the protein structure right from the N-terminal to the C-terminal. This observation is in line with the binding site predictions recorded earlier in the study where residues Thr 91, Phe 92, Phe 193, Asn 279, Gly 283, Ala 284, and Thr 288 of CYP1A1 were highlighted to commonly participate in all the CYP1A1-flavonoid interactions. Moreover, the RoG was evaluated to determine the degree of protein compactness and the stability of the protein during the simulation run. As the compactness of a protein in a complex is induced by the movement of its ligand, higher RoG values indicate less structural compactness and increased conformational entropy, while lower values indicate stable and highly compatible structures [52]. Hence, it can be deduced that the trajectories of the docked complexes remain relatively stable and do not induce any structural changes throughout the simulation indicating appropriate conformation, stability, and condensed structures. Finally, the SASA for the flavonoid-protein complexes was also monitored as an indication of the free energy of each atom’s non-polar solvation in molecules. SASA is an important factor associated with the exposures of hydrophobic residues and solvent receptors during the simulation and it further establishes the stability of docked complexes [52,53]. Furthermore, the closeness in the SASA value exhibited between the apo structure, the standard, silibinin, and the selected P. curatellifolia flavonoids depicts that the binding of the various flavonoids does not alter the enzyme folding. Hence, this simulation study establishes the conformational stability and decent binding potential of P. curatellifolia flavonoids with CYP1A1.
CONCLUSION
The scientific validation of many plant species has demonstrated their significant activities against hepatic damage, thus, the need for further exploration to develop drugs and nutraceuticals from these natural repositories. Hence, the approach in this study was to correlate the flavonoid contents of P. curatellifolia with the previously highlighted hepatoprotective properties of the plant using a combination of computational methods. Hence, structure-based virtual analyses were effectively utilized in this research to further highlight the promising hepatoprotective agents in the plant extract, giving a vivid insight into their bioactivities, safety, and probable mechanisms of action. Subsequently, chrysin, kaempferol, pinocembrin, and quercetin were chosen as the potent lead molecules based on the bioactivity and toxicity prediction as well as preliminary docking experiments. Molecular dynamic simulation of the four selected flavonoids demonstrated promising binding affinities with the target protein, CYP1A1. In addition, post-dynamics analyses of the various CYP1A1-flavonoid complexes were consistent with ligand-enzyme affinity and stability. Results from this study reveal a variety of flavonoid compounds from P. curatellifolia, especially chrysin, kaempferol, pinocembrin, and quercetin, as potential hepatoprotective therapeutics, being found to compare favorably with the standard compound, silibinin. However, there is still the need to investigate the interaction between these flavonoid compounds with other isoforms of the Cytochrome P450 enzyme family as well as with other target proteins involved in xenobiotic metabolism. Furthermore, in vitro and/or in vivo studies of these highlighted flavonoids are recommended as a promising starting point for developing natural anti-hepatotoxic drug candidates. It is believed that results from this study have validated the folkloric use and previous scientific findings on P. curatellifolia and its flavonoid constituents in hepatoprotection, hence, a remarkable foundation has been laid for future exploration.
LIST OF ABBREVIATIONS
3MQ: 3-methylquercetin; CYP1A2: cytochrome P450 family 1 subfamily A member 2; HPLC-DAD: high performance liquid chromatography: diode-array detection; MDS: molecular dynamic simulation; PASS: prediction of activity spectra for substances; RMSD: root mean square deviation; RMSF: root means square fluctuation; RoG: radius of gyration; SASA: solvent accessible surface area; ΔEelec: electrostatic energy; ΔEgas: gas phase free energy; ΔEvdW: van der Waals energy; ΔGbind: total binding free energy; ΔGsolv: solvation free energy.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
FUNDING
There is no funding to report.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
SUPPLEMENTARY DATA
Supplementary data can be found attached to this link here: https://japsonline.com/admin/php/uploadss/4361_pdf.pdf
REFERENCES
1. Olaleye MT, Amobonye AE, Komolafe K, Akinmoladun AC. Protective effects of Parinari curatellifolia flavonoids against acetaminophen-induced hepatic necrosis in rats. Saudi J Biol Sci. 2014;21:486–92. CrossRef
2. Chatepa LEC, Masamba K, Jose M. Proximate composition, physical characteristics and mineral content of fruit, pulp and seeds of Parinari curatellifolia (Maula) from Central Malawi. Afr J Food Sci. 2018;12:238–45. CrossRef
3. Benhura M, Muchuweti M, Gombiro P, Benhura C. Properties of (Parinari curatellifolia) (Hacha or Chakata) fruit from different parts of Harare, Zimbabwe. Afr J Food Agric Nutr Dev. 2013;13(4):8004–18. CrossRef
4. De Wet, Nciki S, van Vuuren SF. Medicinal plants used for the treatment of various skin disorders by a rural community in northern Maputaland. South Afr J Ethnobiol Ethnomed. 2013;9:51. CrossRef
5. Shai KN, Ncama K, Ndhlovu PT, Struwig M, Aremu AO. An exploratory study on the diverse uses and benefits of locally-sourced fruit species in three villages of Mpumalanga Province, South Africa. Foods 2020;9:1581.
6. Halilu EM, October N, Ugwah-Oguejiofor CJ, Jega AY, Nefai, MS. Anti-snake venom and analgesic activities of extracts and betulinic and oleanolic acids isolated from Parinari curatellifolia. J Med Plants Econ Dev. 2020;4:1–8.
7. Manuwa TR, Akinmoladun AC, Crown OO, Komolafe K, Olaleye MT. Toxicological assessment, and ameliorative effects of Parinari curatellifolia alkaloids on triton-induced hyperlipidemia and atherogenicity in rats. Proc Natl Acad Sci, India Sect B Biol Sci. 2017;87:611–23. CrossRef
8. Mawire P, Mozirandi W, Heydenreich M, Chi GF, Mukanganyama S. Isolation and antimicrobial activities of phytochemicals from Parinari curatellifolia (Chrysobalanaceae). Adv Pharmacol Pharm Sci. 2021;2021:1–18. CrossRef
9. Gororo M, Chimponda T, Chirisa E, Mukanganyama S. Multiple cellular effects of leaf extracts from Parinari curatellifolia. BMC Complement Altern Med. 2016;16:1–14. CrossRef
10. Josiah SS, Oyeleye SI, Crown OO, Olaleye MT. Ameliorative effect of Parinari curatellifolia seed extracts on sodium nitroprusside–induced cardiovascular toxicity in rats. Comp Clin Pathol. 2020;29:239 –46. CrossRef
11. Atawodi, S, Yakubu O, Umar I. Antioxidant and hepatoprotective effects of Parinari curatellifolia root. Int J Agric Biol. 2013;15:523–8.
12. Yakubu O, Atawodi S, Ojogbane E, Nwaneri-Chidozie V. Acute toxicity and antioxidant activity of Parinari curatellifollia root methanolic extract in carbon tetrachloride-induced toxicity in wistar rats. Int J Basic Appl Chem Sci. 2012;2:82–9.
13. Marceddu R, Dinolfo L, Carrubba A, Sarno M, Di Miceli G. Milk thistle (Silybum Marianum L.) as a novel multipurpose crop for agriculture in marginal environments: a review. Agronomy. 2022;12:729. CrossRef
14. Boojar MMA, Golmohammad S. Overview of Silibinin anti-tumor effects. J Herb Med. 2020;23:100375. CrossRef
15. Crown OO, Komolafe TR, Akinmoladun AC, Olaleye MT, Akindahunsi AA, Boligon AA. Parinari curatellifolia seed flavonoids protect against triton-induced dyslipidemia and atherogenicity in rats. Trad Kampo Med. 2018;5:11–8. CrossRef
16. Agati G, Brunetti C, Fini A, Gori A, Guidi L, Landi M, et al. Are flavonoids effective antioxidants in plants? Twenty years of our investigation. Antioxidants. 2020;9:1098. CrossRef
17. Mondal S, Rahaman S. Flavonoids: a vital resource in healthcare and medicine. Pharm Pharmacol Int J. 2020;8:91–104. CrossRef
18. Federico A, Dallio M, Loguercio C. Silymarin/silybin and chronic liver disease: a marriage of many years. Molecules. 2017;22:191.
19. RajaSekhar K, RajendraPrasad Y, Shankarananth V, Harika KS, Rajani K, Padmavathamma M. In silico prediction of selected pharmacokinetic, biological and toxic properties of some 1, 3, 5-trisubstituted-2-pyrazolines derived from isonicotinic acid. J Global Trend Pharm Sci. 2011;2:489–512.
20. Evans, W.C. Trease and Evans’ pharmacognosy E-book. Elsevier Health Sciences, Oxford, UK; 2009.
21. Sofowora A. Research on medicinal plants and traditional medicine in Africa. J Altern Complement Med. 1996;2:365–72. CrossRef
22. Öztürk N, Tunçel M, Poto?lu-Erkara ?. Phenolic compounds and antioxidant activities of some Hypericum species: a comparative study with Hypericum perforatum. Pharm Biol. 2009;47:120–7. CrossRef
23. Znati M, Zardi-Bergaoui A, Daami-Remadi M, Ben Jannet H. Semi-synthesis, antibacterial, anticholinesterase activities, and drug likeness properties of new analogues of coumarins isolated from Ferula lutea (Poir.) Maire. Chem Afr 2020;3:635–45. CrossRef
24. Alodeani EA, Arshad M, Izhar MA. Anti-uropathogenic activity, drug likeness, physicochemical and molecular docking assessment of (E-)-N′-(substituted-benzylidene)-2-(quinolin-8-yloxy) acetohydrazide. Asian Pac J Tropical Biomed. 2015;5:676–83. CrossRef
25. Amobonye A, Bhagwat P, Ranjith D, Mohanlall V, Pillai S. Characterisation, pathogenicity and hydrolytic enzyme profiling of selected Fusarium species and their inhibition by novel coumarins. Arch Microbiol. 2021;1–14.
26. Bhagwat P, Amobonye A, Singh S, Pillai S. A comparative analysis of GH18 chitinases and their isoforms from Beauveria bassiana: an in-silico approach. Process Biochem. 2021;100:207–16. CrossRef
27. Shode F, Idowu A, Uhomoibhi O, Sabiu S. Repurposing drugs and identification of inhibitors of integral proteins (spike protein and main protease) of SARS-CoV-2. J Biomol Str Dyn. 2021;1–16. CrossRef
28. Narayanaswamy V, Alaabed S, Obaidat IM. Molecular simulation of adsorption of methylene blue and rhodamine B on graphene and graphene oxide for water purification. Mater Today Proc. 2020;28:1078–83. CrossRef
29. Ibrahim MAA, Abdelrahman AHM, Hussien TA, Badr EAA, Mohamed TA, El-Seedi HR, et al. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput Biol Med. 2020;26:104046.
30. Trivedi A, Ahmad R, Siddiqui S, Misra A, Khan MA, Srivastava A, et al. Prophylactic and therapeutic potential of selected immunomodulatory agents from Ayurveda against coronaviruses amidst the current formidable scenario: an in-silico analysis. J Biomol Str Dyn. 2021;1–53. CrossRef
31. Yu HH, Qiu YX, Li B, Peng CY, Zeng R, Wang W. Kadsura heteroclita stem ethanol extract protects against carbon tetrachloride-induced liver injury in mice via suppression of oxidative stress, inflammation, and apoptosis. J Ethnopharmacol. 2021;267:113496. CrossRef
32. Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. Sci World J. 2013; 2013:162750. CrossRef
33. Shlini, P. Molecular and pharmacokinetic properties of the histidine decarboxylase inhibitors from clove. Int J Green Pharm. 2020;14:203–13. CrossRef
34. Kumar BP, Soni M, Bhikhalal UB, Kakkot IR, Jagadeesh M, Bommu P, et al. Nanjan M. Analysis of physicochemical properties for drugs from nature. Med Chem Res. 2010;19:984–92. CrossRef
35. Sladek FM. What are nuclear receptor ligands? Mol Cell Endocrinol. 2011;334:3–13. CrossRef
36. Zollner G, Trauner M. Nuclear receptors as therapeutic targets in cholestatic liver diseases. Br J Pharmacol. 2009;156:7–27. CrossRef
37. Yang X, Gonzalez FJ, Huang M, Bi H. Nuclear receptors and non-alcoholic fatty liver disease: an update. Liver Res. 2020;4(2):88–93. CrossRef
38. Townsend S, Newsome P. New treatments in non-alcoholic fatty liver disease. Alimentary Pharmacol Ther. 2017;46:494–507. CrossRef
39. Sousa A, Fernandes D, Ferreira M, Cordeiro L, Souza M, Pessoa H, et al. Analysis of the toxicological and pharmacokinetic profile of Kaempferol-3-O-β-D-(6”-Ep-coumaryl) glucopyranoside-tiliroside: in silico, in vitro and ex vivo assay. Braz J Biol. 2021;83:e244127. CrossRef
40. Thorgersen EB, Barratt-Due A, Haugaa H, Harboe M, Pischke SE, Nilsson PH, et al. The role of complement in liver injury, regeneration, and transplantation. Hepatology. 2019;70:725–36. CrossRef
41. Abdel-Salam OM, Youness ER, Mohammed NA, Yassen NN, Khadrawy YA, El-Toukhy SE et al. Nitric oxide synthase inhibitors protect against brain and liver damage caused by acute malathion intoxication. Asian Pac J Tropical Med. 2017;10:773–86. CrossRef
42. Li X, He X, Chen S, Le Y, Bryant MS, Guo L, et al. The genotoxicity potential of luteolin is enhanced by CYP1A1 and CYP1A2 in human lymphoblastoid TK6 cells. Toxicol Lett. 2021;344:58–68. CrossRef
43. Salgueiro AC, Folmer V, da Rosa HS, Costa MT, Boligon AA, Paula FR, et al. In vitro and in silico antioxidant and toxicological activities of Achyrocline satureioides. J Ethnopharmacol. 2016;194:6–14. CrossRef
44. Stading R, Couroucli X, Lingappan K, Moorthy B. The role of cytochrome P450 (CYP) enzymes in hyperoxic lung injury. Expert Opin Drug Metab Toxicol. 2021;17:171–8. CrossRef
45. Uno S, Nebert DW, Makishima M. Cytochrome P450 1A1 (CYP1A1) protects against nonalcoholic fatty liver disease caused by western diet containing benzo[a]pyrene in mice. Food Chem Toxicol. 2018;113:73–82. CrossRef
46. Coelho AM, Queiroz IF, Lima WG, Talvani A, Perucci LO, Oliveira de Souza M, et al. Temporal analysis of paracetamol-induced hepatotoxicity. Drug Chem. Toxicol. 2022; 1–10.
47. Salmaso V, Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front Pharmacol. 2018;9:923. CrossRef
48. Fang J, Wu P, Yang R, Gao L, Li C, Wang D, et al. Inhibition of acetylcholinesterase by two genistein derivatives: kinetic analysis, molecular docking and molecular dynamics simulation. Acta Pharm Sinica B. 2014;4:430–37. CrossRef
49. Hejazi II, Beg MA, Imam MA, Athar F, Islam A. Glossary of phytoconstituents: can these be repurposed against SARS CoV-2? A quick in silico screening of various phytoconstituents from plant Glycyrrhiza glabra with SARS CoV-2 main protease. Food Chem Toxicol. 2021;150:112057. CrossRef
50. Naidoo D, Roy A, Kar P, Mutanda T, Anandraj A. Cyanobacterial metabolites as promising drug leads against the Mpro and PLpro of SARS-CoV-2: An in silico analysis. J Biomol Str Dyn. 2020;1–13. CrossRef
51. Muthumanickam S, Indhumathi T, Boomi P, Balajee R, Jeyakanthan J, Anand K, et al. In silico approach of naringin as potent phosphatase and tensin homolog (PTEN) protein agonist against prostate cancer. J Biomol Str Dyn. 2020;1–10. CrossRef
52. Singh G, Tiwari A, Choudhir G, Kumar A, Sharma P. Unravelling the potential role of bioactive molecules produced by Trichoderma spp. as inhibitors of tomatinase enzyme having an important role in wilting disease: an in-silico approach. J Biomol Str Dyn. 2021;1–10. CrossRef
53. Mishra A, Pathak Y, Kumar A, Mishra SK, Tripathi V. Natural compounds as potential inhibitors of SARS-CoV-2 main protease: An in-silico study. Asian Pac J Trop Biomed. 2021;11:155–63. CrossRef