INTRODUCTION
Marine resources have become the center of new bioactive mining in recent decades [1–3]. The unique environmental condition in marine ecosystems drives the organisms to produce a wide range of secondary metabolites to protect themselves from unfavorable conditions. Plenty of secondary metabolites from marine organisms were reported as novel compounds with prospective biological activities for pharmaceutical and medicinal applications [4–6]. For instance, the food and drug administration (FDA) has approved several marine natural products for medicinal purposes, such as cytarabine, vidarabine, trabectedin, and ziconotide [7]. This robust prospect triggers more investigation of bioactive compounds from marine organisms.
As a tropical and maritime country, Indonesia stashes massive marine bioresources with remarkable biological activities [8]. In addition, most marine natural products from Indonesia were regularly reported from Porifera and Cnidarian [9,10]. Interestingly, Cnidarian-originated natural products are mostly isolated from soft corals, while stony corals are rarely reported [11,12]. It is suggested due to the low amount of soft tissues that could be extracted and the lack of ability to produce novel compounds. Regardless of this condition, stony corals harbor a wide variety of species of bacteria, fungi, and other microorganisms [13,14]. Coral-associated bacteria received massive attention as the source of potentially bioactive compounds due to their ability to produce secondary metabolites [15–18].
It is noted that actinobacteria produce more diverse and interesting bioactive compounds among other groups of coral-associated bacteria. In addition, Streptomyces, Micromonospora, and Nocardiopsis are the three genera of actinobacteria that have been repeatedly reported as the most potential producers of secondary metabolites from the marine environment [19]. Our previous works have discovered novel compounds from stony coral-associated-actinobacteria: labrenzbactin, nocarimidazoles C-D, and (2Z,4E)-3-methyl-2,4-decadienoic acid. These natural products exhibited bacterial and cytotoxic properties [15,16,20,21]. In addition, iseolides A-C were isolated from coral-associated Streptomyces sp. with antifungal activity [18].
Their ability to produce bioactive compounds leads our current study to explore indigenous coral-associated actinobacteria from Indonesia. As a part of the world’s coral triangle, Indonesia harbors various untapped coral-associated actinobacteria that potentially produce interesting secondary metabolites [22]. Therefore, this study was conducted to isolate and identify coral-associated actinobacteria from Karimunjawa National Park, Indonesia; to investigate their antimicrobial and cytotoxic properties; to acquire the most potent actinobacteria with its suitable media for bioactive productions; and to characterize the secondary metabolites from the prospective extracts using high-performance liquid chromatography with diode array detector (HPLC-DAD)-ultraviolet (UV)-Vis guided analysis.
MATERIALS AND METHODS
Sampling
A solitary stony coral was collected from Menjangan Besar Island, Karimunjawa National Park, Jepara region, Indonesia, at a depth of 15 m by SCUBA diving (Fig. 1). The sample was kept inside a sterile ziplock plastic and then transferred to a cold box for preservation. This step was conducted aseptically to prevent cross-contamination from the environment [23–25].
Bacterial isolation, purification and identification
Bacterial isolation was conducted within 5 hours of sample collection to prevent the alteration of microbial abundance from the sample. Serial dilution method was performed in this study. ISP4 agar (DifcoTM) with the addition of humus 1% (purchased from a local agricultural store in Semarang) was applied for bacterial isolation. Two agar plates were opened during bacterial isolation as environmental control. After 2–4 weeks of incubation period, each bacterium on the isolation plate with different colony morphology from the bacteria growing on the environmental control plate was purified onto ISP 4 agar. Afterward, a molecular study based on 16S rRNA gene sequence was carried out to identify each bacterium. The protocols for DNA extraction to amplification steps were conducted following our previous work [25,26]. The polymerase chain reaction products were sent to the 1st Base Laboratories Sdn Bhd, Malaysia, for DNA sequencing. Each sequence was analyzed based on its homology on a Basic Local Alignment Search Tool. The phylogenetic tree was reconstructed using the maximum-likelihood method on Molecular Evolutionary Genetics Analysis 11 software. Furthermore, only actinobacteria isolates were selected for further experiments.
Metabolites production and extraction
All actinobacteria strains were cultivated in V22 medium (1% soluble starch, 0.5% glucose, 0.3% NZ-caze, 0.2% yeast extract, 0.5% Tryptone, 0.1% K2HPO4, 0.005% MgSO4.7H2O, 0.3% CaCO3, natural seawater—final pH 7) as a seed culture then incubated for four days at 30°C (200 r.p.m.). Then, seed cultures were transferred into three different media namely A3 (2% soluble starch, 0.5% glucose, 2% glycerol, 0.3% yeast extract, 1.5% Pharmamedia, and 1% Diaion HP-20, natural seawater—final pH 7), A11 (2% glucose, 2.5% soluble starch, 0.5% yeast extract, 0.5% polypeptone, 0.5% NZ-amine, 0.5% CaCO3, 1% Diaion HP-20, natural seawater—final pH 7), and A16 (2% glucose, 1% Pharmamedia, 0.5% CaCO3, 1% Diaion HP-20 natural seawater—final pH 7) in a K-1 flask. The production culture was incubated using a similar condition as the seed culture [27,28]. At the end of the incubation session, the samples were extracted using 1-butanol. The crude extracts were concentrated using a rotary evaporator and stored at −20°C for subsequent analysis [25].
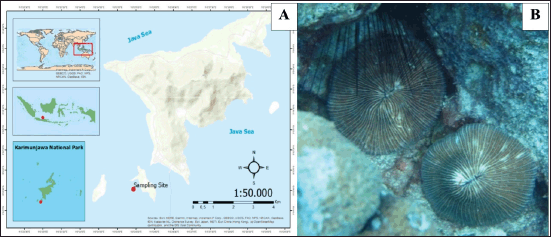 | Figure 1. (A) Sampling site at Menjangan Besar Island, Karimunjawa National Park. (B) In situ photograph of Fungia sp. [Click here to view] |
Antimicrobial assay
A paper disc diffusion method was conducted as described by the Clinical and Laboratory Standards Institute [29] with several modifications. Escherichia coli NIHJ JC-2, Micrococcus luteus ATCC9341, Staphylococcus aureus FDA209P JC-1, and Candida albicans NBRC0197 were the pathogen collections in Toyama Prefectural University, Japan. The bacterial pathogens were cultivated on Mueller Hinton agar (MHA), while fungal pathogen on sabouraud dextrose agar. Each crude extract was diluted in dimethyl sulfoxide (DMSO) to reach a concentration of 500 μg/ml. A total of 10 μl of crude extract was injected into the paper disc, then transferred onto an agar plate that had been inoculated with the tested pathogen. The plates were incubated at 37°C for 24 hours. A positive result was indicated by the presence of an inhibition zone around the paper disc. Furthermore, the active extracts were continued to determine the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values.
Determination of MIC and MBC values was conducted according to Balouiri et al. [30] using the 96-well plate broth microdilution method. The crude extracts were diluted in DMSO. The bacterial pathogens were cultivated in Mueller Hinton Broth (MHB) to reach a density of 0.5 McFarland. A total of 100 μl broth media was transferred into each well. Then, 90 μl of pathogen culture and 10 μl of extract solution were transferred into the first well to reach a total volume of 200 μl. Afterward, a serial dilution was performed; therefore, 100 μl mixture remained in each well. Consequently, 100 μl of MHB was added to each well to make a final volume of 200 μl/well. Hence, the concentration range was 500 –1.95 μg/ml. The plate was incubated at 37°C for 24 hours. Thereafter, 20 μl of WST-1 (Roche) was added into each well. The plate was reincubated for 2 hours. A deep yellow to orange color indicated bacterial growth, while a colorless indicated the inhibition of bacterial growth. The lowest concentration, which able to inhibit bacterial growth, was chosen as the MIC value.
A total of 100 μl of bacterial culture from the well that inhibits bacterial growth was inoculated onto the MHA plate, then incubated for 24 hours. The concentration with no bacterial growth on the plate was determined as the MBC value.
Cytotoxicity assay against P388 murine leukemia cancer cell
All actinobacterial extracts were diluted in DMSO for the screening of cytotoxic assay with a concentration of 1 mg/ml. The cells were treated with the diluted extracts: DMSO as the negative control and doxorubicin as the positive control. The plates were incubated for 72 hours at 37°C with 5% CO2 atmosphere in the air and 100% humidity. Subsequently, 50 μl of solution containing XTT (1 mg/ml) and PMS (40 μg/ml) was transferred into each well and reincubated for 4 hours under similar conditions. The cell viability percentage was observed using a microplate reader with a 450 nm absorbance wavelength. The % of cell viability was calculated by the following formula:
The calculation of the IC50 value was merely applied for the prospective crude extracts with <40% cell viability. Each promising crude extract was diluted using DMSO into five concentrations, such as 0.0003, 0.003, 0.03, 0.3, and 3 mg/ml. Then the same condition as screening was applied to culture the cells [15,16,25]. Three replications were applied in this assay.
Metabolite characterization of prospective extracts
HPLC-DAD-UV-Vis was applied to characterize the bioactive compounds from the prospective crude extracts with antimicrobial and cytotoxic activity. 1 mg/ml of crude extract was injected into HPLC-DAD-UV-Vis with acetonitrile (Wako, Japan) and 0.1% formic acid (Wako, Japan) solution as the eluent system, whereas COSMOCIL 3C18-AR-II (4.6ID × 100 mm) from Nacalai Tesque as the column. The flow rate was 1.2 ml/minute, and 160 bar in pressure with this following condition: 0%–40% of acetonitrile for 0–25 minutes, 40%–85% for 25–28 minutes, 85% for 28–30 minutes, and 85%–90% for 30–35 minutes. The pattern of UV spectrum, UV λmax, and retention time (RT) of each peak was compared to the in-house actinobacteria metabolites database [27,28,31,32].
Data analysis
The IC50 value for cytotoxicity was calculated by plotting the triplicate data to a single-logarithmic chart in MS Excel [15,16]. Then, the data were analyzed using SPSS package with confidence interval of 95% (p < 0.05).
RESULTS AND DISCUSSION
Composition of Fungia sp. associated bacteria
Karimunjawa National Park is one of the oldest marine conservation areas in Indonesia. This national park consists of 27 islands with three classified zones: marine protection area (MPA) zone, utilization zone, and non-marine protection (non-MPA) zone. Moreover, Menjangan Besar Island is part of the utilization zone for tourism [33]. From this location, a solitary unidentified stony coral was collected with the following morphological features: a brown color, circular polyp with a central arch, diameter of 16 cm, monostomatous, mouth located in the center, and free-living (nonattached) (Fig. 1B). These characteristics belong to genus Fungia. This genus is a solitary stony coral commonly found in tropical countries, including Indonesia [34]. A prior study by Kennedy et al. [33] stated that Fungia is one of the stony coral genera that live in Menjangan Island, Karimunjawa National Park, with a low percentage.
Corals harbor numerous microorganisms, including bacteria, in their mucus, tissue, and calcium carbonate skeleton. It is highlighted that bacteria play an important role in nutrient cycling and maintaining coral’s health and resilience [14]. In addition, their ability to produce particular chemical substances to protect their host from pathogens and other creatures [35] leads our current study to work with a stony coral, Fungia sp. Interestingly, the composition of associated bacteria from this stony coral is less reported rather than in other genera [12,17,36]. Even this study is the first report of Fungia-associated bacteria from Indonesia.
 | Figure 2. Phylogenetic tree of Fungi sp. associated bacterium based on 16S rRNA gene sequence analysis. [Click here to view] |
In total, 13 bacterial colonies with distinct morphological features were obtained from the sample. The 16S rRNA gene sequence revealed that these 13 species belonged to 10 genera and 3 phyla (Fig. 2). The most abundant phylum was proteobacteria (53.84%), followed by actinobacteria (30.76%) and firmicutes (15.40%). Some studies which applied culture-independent approach to investigate bacterial diversity in stony corals stated that proteobacteria and firmicutes were noted as the common dominant phylum, while actinobacteria as a minor phylum [37,38]. Our result was different because this study applied a culture-dependent method with ISP 4 agar as the culture medium. This medium contains suitable nutrients to grow actinobacteria, especially Streptomyces. The absence of antibiotics such as nalidixic acid in agar medium for bacterial isolation is suspected to cause non-actinobacterial growth [39,40].
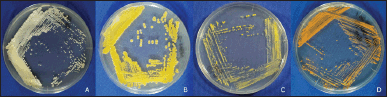 | Figure 3. Colonial morphology of (A) Streptomyces pluripotens CM4, (B) Streptomyces ardesiacus CM11, (C) Micrococcus flavus CM 13, and (D) Gordonia hongkongensis CM20. [Click here to view] |
Among the 13 bacteria, it was highlighted that four of them were identified as actinobacteria, namely, Streptomyces pluripotens CM4, Streptomyces ardesiacus CM11, Micrococcus flavus CM13, and Gordonia hongkongensis CM20 (Fig. 3). Previously, genera Streptomyces, Micrococcus, and Gordonia were also reported as coral-associated bacteria [25,38,41]. As it has been mentioned before, this study merely focused on actinobacteria because the members of this phylum are repeatedly reported to produce remarkable bioactive compounds especially antimicrobial and anticancer [17,19,42]. Therefore, only these four isolates were continued to the further steps.
Antibacterial activity
Four actinobacteria strains were cultivated in three different media for antimicrobial compound production. Table 1 and Figure 4 show the result of antimicrobial assay. Some extracts from S. pluripotens CM4 and S. ardesiacus CM11 were noted to have antibacterial properties. It has been known that most antibiotics in the market nowadays were discovered from terrestrial Streptomyces [43]. Therefore, our finding is very important for the discovery of future antibiotic compounds from marine Streptomyces. The crude extract of S. pluripotens CM4 cultivated in A3 and A11 media only inhibited the growth of M. luteus. On the other hand, S. ardesiacus CM11, which was cultivated in A11 medium, was highlighted as the most potential extract due to its ability to inhibit all pathogenic bacteria in this study, followed by the extract of A3 medium, which successfully inhibited M. luteus and S. aureus, then extract from A16 medium which only inhibited M. luteus. However, none of the extracts inhibit the growth of C. albicans.
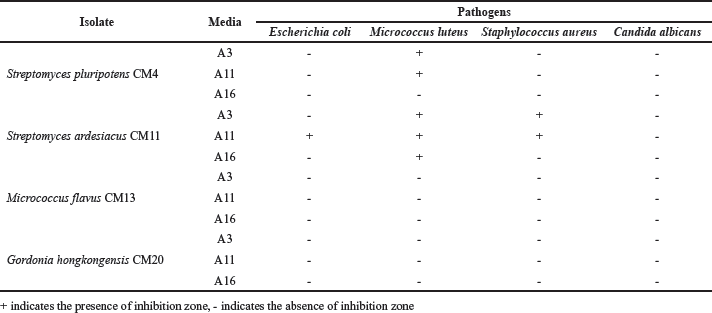 | Table 1. Antimicrobial activity of Fungia sp.-associated actinobacteria using paper disc diffusion method. [Click here to view] |
 | Figure 4. Inhibition zones of some prospectives bacterial extracts against (A) M. luteus, (B) E. coli, and (C) S. aureus. [(A.1) CM4 cultivated in medium A3. (A.2) CM4 cultivated in medium A11. (A.3, B, and C.1) CM11 cultivated in medium A3. (A.4 and C.2) CM11 cultivated in medium A11. (A.4) CM11 cultivated in medium A16.]. [Click here to view] |
Then, these prospective extracts proceeded for MIC and MBC values determination. The result of this step is presented in Table 2. We noted that the extract of S. ardesiacus CM11, which was cultivated in an A11 medium, exhibited the lowest MIC and MBC against all bacterial pathogens with a range of MIC value of 7.81–31.25 μg/ml, while the MBC range was 7.81–62.50 μg/ml. On the other hand, extracts from S. pluripotens CM4 had a MIC value range of 15.62–31.25 μg/ml, whereas the range of MBC value was 15.62–62.50 μg/ml. Broth microdilution is a common method to determine MIC and MBC value because it can save media, reagents, and the substances that need to be tested. Moreover, a lower MIC value means the substances have better antimicrobial activity [44]. Moreover, this method is also suggested by the CLSI as a standard protocol to conduct antimicrobial susceptibility tests on pathogens [29].
According to its MIC and MBC value, it was suggested that the A11 medium provided better nutrition for S. pluripotens CM4 and S. ardesiacus CM11 to produce antimicrobial compounds. Our previous study also applied this medium to produce several new antibacterial compounds from coral-associated bacteria, such as nocarimidazoles C-D [15,16]. Besides, this medium was also chosen to produce antimicrobial compounds from actinobacteria such as TMKS8A from Streptomyces sp. TMKS8 [32] and nomimicins B-D from Actinomadura sp. [28].
Cytotoxicity
Cytotoxicity is defined as the toxic effect of bioactive compounds on the targeted living cells. This assay is commonly applied to screen for anticancer properties of bioactive substances [45]. Cancer is a disease triggered by abnormal and uncontrollable cell growth in the body and then invades other body parts. WHO, through The Global Cancer Observatory, released global data on cancer. It was recorded that in 2020, there were more than 19.2 mil new cancer cases, and more than 9.9 mil of cases led to death. Leukemia was noted as one of the leading cancers that contributed to high mortality of the patient, with a number of deaths of more than 300,000 cases only in 2020 globally [46]. In Indonesia, leukemia is ranked 9th as the deadliest cancer, killing more than 11,000 people in 2020 [47]. Unfortunately, some drug resistance was also reported in many leukemia cases worldwide [48–50]. Therefore, finding a novel anticancer agent to treat leukemia is urgent. Thus, this study conducted a cytotoxicity assay to combat leukemia cancer cells. The result of this assay is shown in Fig. 5.
Among all isolates, two extracts of S. pluripotens CM4 and three extracts of S. ardesiacus CM11 showed a potential anticancer activity with cell viability < 40%. The crude extract killed > 60% of the tested cancer cells. Statistically, Table 3 shows that the extract of S. ardesiacus CM11 from the A3 medium had the lowest IC50 value (4.43 ± 2.85 μg/ml), while the highest IC50 value belonged to S. pluripotens CM4 extract from A11 medium (26.54 ± 1.03 μg/ml).
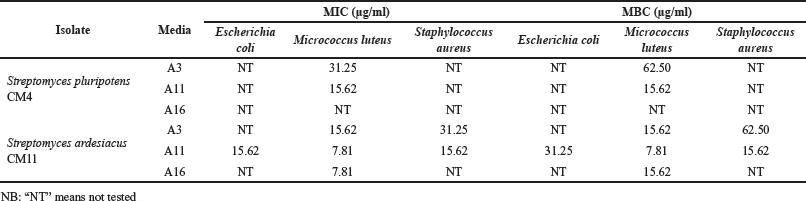 | Table 2. MIC and MBC values of prospective Fungia sp.-associated actinobacteria. [Click here to view] |
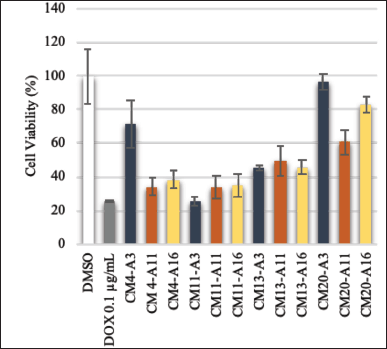 | Figure 5. Cell viability of P388 Murine leukemia cancer cells after treated with bacterial crude extracts with concentration of 1 mg/ml. Doxorubicin (Dox) as control positive and DMSO as control negative. [Click here to view] |
IC50 is a value to indicate the concentration of a particular substance to kill 50% of the tested cells. Therefore, a lower IC50 value means the substance is more efficient to kill the tested cells [51]. Hence, S. ardesiacus CM11 extract from the A3 medium was considered to has the best anticancer potential among other extracts. In addition, a sponge-associated S. ardesiacus has been reported previously as a producer of antibacterial and anticancer compounds, namely, urdamycins W-X and grincamycin U [52].
Furthermore, our study indicated that the A3 medium provided better nutrition for S. ardesiacus CM11 to produce anti-cancer compounds. Sharma et al. [21] also used an A3 medium to produce a new compound, namely, labrenzbactin, from a coral-associated bacterium Labrenzia sp. This compound had an IC50 value of 13 mM against P388 Murine Leukemia cells. Another study successfully isolated several new anticancer compounds, namely, pseudosporamide and pseudosporamicins A-C, from a rare genus actinobacteria Pseudosporangium sp. with this medium [27].
 | Table 3. IC50 value of prospective extracts from Fungia sp. associated actinobacteria against P388 Murine leukemia cancer cells. [Click here to view] |
Metabolites profile of prospective extracts
The result of antimicrobial and cytotoxicity assays led to five prospective extracts for metabolite characterization using the HPLC-DAD-UV-Vis guided method. This methodology has been applied to isolate plenty of novel compounds from actinobacteria [18,20,21,27,31,32]. It is noted that the extract of S. pluripotens CM4, which were cultivated in A3 and A11, showed one major peak at RT of 24.39 minutes with a UV pattern as shown by Figure 6A. This peak had UV λmax at 239 nm. According to its UV pattern, UV λmax, and RT, the actinobacteria metabolites database suggested this peak as a member of piericidins derivatives. Piericidins are a specialized metabolite that is merely produced by actinobacteria, especially Streptomyces. In addition, members of piericidins are known to have UV λmax at 239 nm [53]. Hence, this data strengthens our result of metabolites profiling using the HPLC-DAD-UV-Vis method.
Piericidins derivatives are outstanding bioactive compounds with diverse biological activities, especially as antimicrobial and anticancer agents. Moreover, they are known as a broad-spectrum antimicrobial agent [53,54]. In addition, two antimicrobial mechanisms have been identified from piericidins such as by inhibiting the quorum-sensing in some bacteria and blocking the type III secretion system for needle assembly in Yersinia pseudotuberculosis [55–57]. As an anticancer agent, piericidins derivatives had an ability to inhibit several types of human cancer cells, such as renal tumor OS-RC-2, renal carcinoma ACHN, leukemia HL-60, and leukemia K-562 [58]. The presence of piericidins derivatives was expected as the reason for antimicrobial and anticancer properties in S. pluripotens CM4 extracts.
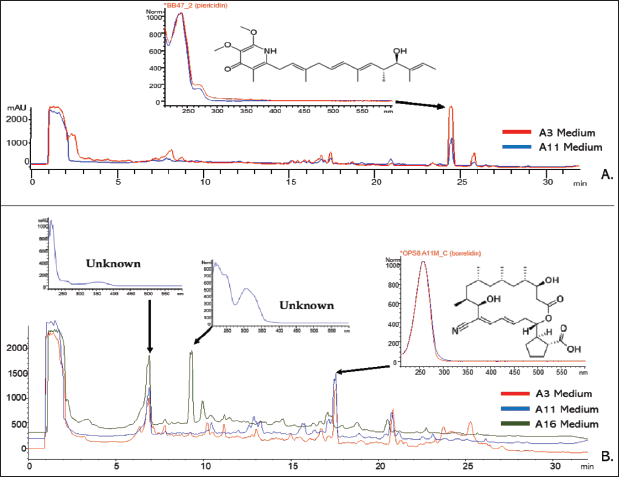 | Figure 6. Chromatogram profile of prospective extracts based on HPLC-DAD-UV-Vis analyses. (A) S. pluripotens CM4. (B) S. ardesiacus CM11). [Click here to view] |
On the other hand, the HPLC-DAD-UV-Vis analysis discovered that S. ardesiacus CM11 produced more compounds which indicated by the number of peaks in HPLC chromatograms. It was noted that all crude extracts from S. ardesiacus CM11 had two major peaks at RT 6.80 minutes with λmax of 221 nm and RT 17.40 minutes with λmax of 257 nm. Then, extract from the A16 medium had another major peak at RT 9.21 minutes with λmax at 217 nm. Unfortunately, among these three major peaks, the in-house actinobacteria metabolite database can merely characterize one peak at RT 17.40 minutes. According to its RT, UV pattern, and λmax, this peak was suggested as borrelidin derivatives (Fig. 6B). Borrelidin is an 18-membered macrolide polyketide characterized by unusual cyclopentane dicarboxylic starter acid and the nitrile functional group [59].
Borrelidins has UV λmax at 257 nm and are mainly produced by Streptomyces spp. This metabolite possesses various biological activity, including antimicrobial, anticancer, and antimalarial [59-61]. Therefore, borrelidin was suggested to be one of the metabolites produced by S. ardesiacus CM11.
It has been known that borrelidin selectively inhibits the threonyl-tRNA synthetase in several species of bacteria and human cells during protein synthesis to kill the cells [61]. In consequence, it has a very outstanding activity as antimicrobial and anticancer agents [62]. In addition, Habibi et al. [63] stated that this compound was able to induce apoptosis in acute lymphoblastic leukemia. Hence, the presence of borrelidin was expected to contribute to the antibacterial and anticancer properties of S. ardesiacus CM11 crude extracts.
Although the result of HPLC-DAD-UV-Vis analysis successfully identified two peaks from our samples as piericidins and borrelidins derivatives, it is highly recommended to carry out a further analysis on the basis of molecular weight to confirm the exact compounds. Even though marine Streptomyces produce enormous number of known compounds, the discovery of novel metabolites from this genus is still regularly reported [43,64]. It means that our strains also have potential to produce novel compounds. In addition, our previous works effectively isolated many new compounds from actinobacteria, non-actinobacteria, and fungi based on UV pattern and λmax using HPLC-DAD-UV-Vis guided method [15,16,18,20,21,26,31,32]. Therefore, the unidentified major peaks in the crude extract were expected as the candidates of novel compounds for our future project.
CONCLUSION
Our work successfully isolated 13 associated bacteria from Fungia sp. Among all isolates, four strains were identified as actinobacteria based on their 16S rRNA gene sequence. The result of bioassays revealed that the crude extract of S. ardesiacus CM11 from A11 medium exhibited the lowest MIC (7.81–31.25 μg/ml) and MBC (7.81–62.50 μg/ml) value against all bacterial pathogens. Furthermore, crude extract of S. ardesiacus CM11 from A11 medium, exhibited the strongest cytotoxicity against P388 Murine Leukemia Cancer Cells with an IC50 value of 4.43 ± 2.85 μg/ml.
ACKNOWLEDGMENTS
The authors also thank Balai Taman Nasional Karimunjawa (BTNK) for granting us a sampling permit.
FINANCIAL SUPPORT
The authors gratefully acknowledge Universitas Diponegoro for funding this research through Riset Publikasi Internasional Bereputasi Tinggi (RPIBT) scheme with contract number 185-96/UN7.6.1/PP/2022. This work was also partially funded by Toyama Prefectural University, Japan, through Special Research Fellowship Grant granted to the first author.
CONFLICT OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
DATA AVAILABILITY
All data generated and analyzed are included in this research article.
PUBLISHER’S NOTE
This journal remains neutral with aregard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Pawlik JR. Handbook of marine natural products. Springer; 2012. pp 677–710.
2. Blunt JW, Caroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep. 2018;35(1):8–53.
3. Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep. 2019;36(1):122–73.
4. Jime?nez C. Marine natural products in medicinal chemistry. ACS Med Chem Lett. 2018;9(10):959–61.
5. Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep. 2021;38(2):362–413.
6. Carroll AR, Copp BR, Davis RA, Keyzers RA, Prinsep MR. Marine natural products. Nat Prod Rep. 2022;39(6):1122–71.
7. Pereira F. Have marine natural product drug discovery efforts been productive and how can we improve their efficiency? Expert Opin Drug Discov. 2019;14(8):717–22.
8. Putra MY, Murniasih T. Distribution and diversity of marine natural products from Indonesian marine organisms. J Coast Life Med. 2016;4(2):104–7.
9. Hanif N, Murni A, Tanaka C, Tanaka J. Marine natural products from Indonesian waters. Mar Drugs. 2019;17(6):364.
10. Izzati F, Warsito MF, Bayu A, Prasetyoputri A, Atikana A, Sukmarini L, et al. Chemical diversity and biological activity of secondary metabolites isolated from Indonesian marine invertebrates. Molecules. 2021;26(7):1898.
11. Putra MY, Wibowo JT, Murniasih T. A review of chemistry and biological activities of the Indonesian Octocorallia. J App Pharm Sci. 2017;7(5):219–27.
12. Sang VT, Dat TTH, Vinh LB, Cuong LCV, Oanh PTT, Ha H, et al. Coral and coral-associated microorganisms: a prolific source of potential bioactive natural products. Mar Drugs. 2019;17(8):468.
13. Glasl B, Herndl GJ, Frade PR. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016;10(9):2280–92.
14. Sun W, Anbuchezhian R, Li Z. Association of coral-microbes, and the ecological roles of microbial symbionts in corals. In The Cnidaria, past, present and future. Springer; 2016. pp 347–57.
15. Karim MRU, Harunari E, Oku N, Akasaka K, Igarashi Y. Bulbimidazoles AC, antimicrobial and cytotoxic alkanoyl imidazoles from a marine gammaproteobacterium Microbulbifer species. J Nat Prod. 2020;83(4):1295–9.
16. Karim MRU, Harunari E, Sharma AR, Oku N, Akasaka K, Urabe D, et al. Nocarimidazoles C and D, antimicrobial alkanoylimidazoles from a coral-derived actinomycete Kocuria sp.: application of 1JC,H coupling constants for the unequivocal determination of substituted imidazoles and stereochemical diversity of anteisoalkyl chain. Beilstein J Org Chem. 2020;16:2719–27.
17. Mahmoud HM, Kalendar AA. Coral-associated actinobacteria: diversity, abundance, and biotechnological potentials. Front Microbiol. 2016;7:1–13.
18. Zhang Z, Zhou T, Harunari E, Oku N, Igarashi Y. Iseolides A–C, antifungal macrolides from a coral-derived actinomycete of the genus Streptomyces. J Antibiot. 2020;73(8):534–41.
19. Chen J, Xu L, Zhou Y, Han B. Natural products from actinomycetes associated with marine organisms. Mar Drugs. 2021;19(11):629.
20. Sharma AR1, Harunari E, Zhou T, Trianto A, Igarashi Y. Isolation and biosynthesis of an unsaturated fatty acid with unusual methylation pattern from a coral-associated bacterium Microbulbifer sp. Beilstein J Org Chem. 2019;15:2327–32.
21. Sharma AR2, Zhou T, Harunari E, Oku N, Trianto A, Igarashi Y. Labrenzbactin from a coral-associated bacterium Labrenzia sp. J Antibiot. 2019;72(8):634–9.
22. Tanaka J. How can we develop marine natural products chemistry in Indonesia?. IOP Conf Ser J Physic Ser. 2020;1460:012079.
23. Sabdaningsih A, Cristianawati O, Sibero MT, Nuryadi H, Radjasa OK, Sabdono A, et al. Screening antibacterial agent from crude extract of marine-derived fungi associated with soft corals against MDR-Staphylococcus haemolyticus. IOP Conf Ser Earth Environ Sci. 2017;55:012026.
24. Sibero MT1, Bachtiarini TU, Trianto A, Lupita AH, Sari DP, Igarashi Y, et al. Characterization of a yellow pigmented coral-associated bacterium exhibiting anti-bacterial activity against multidrug resistant (MDR) organism. Egyptian J Aqua Res. 2019;45(1):81–7.
25. Sibero MT, Frederick EH, Lubis K, Wijayanti DP, Haryanti D, Siswanto AP, et al. Antibacterial, cytotoxicity, and biosynthetic gene cluster analysis of coral-associated actinobacteria from Menjangan Besar Island, Karimunjawa National Park. Rasayan J Chem. 2022;15(04):2692–702.
26. Sibero MT2, Zhou T, Fukaya K, Urabe D, Radjasa OKK, Sabdono A, et al. Two new aromatic polyketides from a sponge-derived Fusarium. Beilstein J Org Chem. 2019;15:2941–7.
27. Saito S, Atsumi K, Zhou T, Fukaya K, Urabe D, Oku N, et al. A cyclopeptide and three oligomycin-class polyketides produced by an underexplored actinomycete of the genus Pseudosporangium. Beilstein J Org Chem. 2020;16(1):1100–10.
28. Zhang Z2, Sibero MT, Kai A, Fukaya K, Urabe D, Igarashi Y. TMKS8A, an antibacterial and cytotoxic chlorinated α-lapachone, from a sea slug-derived actinomycete of the genus Streptomyces. J Antibiot. 2021;74(7):464–9.
29. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 30th ed. Clinical and Laboratory Standards Institute; 2020.
30. Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: a review. J Pharma Anal. 2016;6(2):71–9.
31. Saito S, Indo K, Oku N, Komaki H, Kawasaki M, Igarashi Y. Unsaturated fatty acids and a prenylated tryptophan derivative from a rare actinomycete of the genus Couchioplanes. Beilstein J Org Chem. 2021;17:2939–49.
32. Zhang Z, Zhou T, Yang T, Fukaya K, Harunari E, Saito S, et al. Nomimicins B-D, new tetronate-class polyketides from a marine-derived actinomycete of the genus Actinomadura. Beilstein J Org Chem. 2021;17:2194–2202.
33. Kennedy EV, Vercelloni J, Neal BP, Ambariyanto, Bryant DEP, Ganasa A, et al. Coral reef community changes in Karimunjawa national park, Indonesia: assessing the efficacy of management in the face of local and global stressors. J Mar Sci Eng. 2020;8(10):1–27.
34. Veron JEN, Stafford-Smith MG, Turak E, DeVantier LM. Coral of the world. 2016. Available from: http://www.coralsoftheworld.org/species_factsheets/?version=0.01
35. Raina JB, Tapiolas D, Motti CA, Foret S, Seemann T, Tebben J, et al. Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. Peer J. 2016;(8)4:e2275.
36. Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc Biol Sci. 2013;280(1755):20122328.
37. Huggett MJ, Apprill A. Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ Microbiol Rep. 2019;11(3):372–85.
38. Vilela CLS, Villela HDM, Rachid CTCC, Carmo FL, Vermelho AB, Peixoto RS. Exploring the diversity and biotechnological potential of cultured and uncultured coral-associated bacteria. Microorganisms. 2021;9(11):2235.
39. Majithiya VR, Gohel SD. Isolation and characterization of marine actinobacteria associated with the seaweeds, Codium dwarkense and Sargassum cinereum, collected from the Veraval coastline, Gujarat, India. J Mar Biol Assoc India. 2022;64(1):33–7.
40. Rathore DS. Isolation strategies, abundance and characteristics of the marine actinomycetes of Kachhighadi, Gujarat, India. J Mar Biol Assoc India. 2019;61(1):71–8.
41. Sun W, Peng C, Zhao Y, Li Z. Functional gene-guided discovery of type II polyketides from culturable actinomycetes associated with soft coral Scleronephthya sp. PLoS One. 2012;7(8):e42847.
42. Anandan R, Dharumadurai D, Manogaran GP. An introduction to actinobacteria. In Actinobacteria. Basics and biotechnological applications. London, UK: IntechOpen; 2016. pp 3–38.
43. Procópio REL, Silva IR, Martins MK, Azevedo JL, Araújo JM. Antibiotics produced by Streptomyces. Braz J Infect Dis. 2012;16(5):466–71.
44. Veiga A, Toledo MGT, Rossa LS, Mengarda M, Stofella NCF, Oliveira LJ, et al. Colorimetric microdilution assay: validation of a standard method for determination of MIC, IC50%, and IC90% of antimicrobial compounds. J Microbiol Methods. 2019;162:50–61.
45. Mukherjee PK. Bioassay-guided isolation and evaluation of herbal drugs. Quality control and evaluation of herbal drugs. Amsterdam, The Netherlands: Elsevier; 2017. pp 515–37.
46. The Global Cancer Observatory. Leukaemia. 2020 [cited 2023 Feb 20]. https://gco.iarc.fr/today/data/factsheets/cancers/36-Leukaemia-fact-sheet.pdf
47. The Global Cancer Observatory. Indonesia. 2021 [cited 2023 Feb 20]. https://gco.iarc.fr/today/data/factsheets/populations/360-indonesia-fact-sheets.pdf.
48. Kaspers GJL, Veerman AJP. Clinical significance of cellular drug resistant in childhood leukemia. Chemosens Test Oncol. 2016;196–220.
49. Choi S, Henderson MJ, Kwan E, Beesley AH, Sutton A, Bahar AY, et al. Relapse in children with acute lymphoblastic leukemia involving selection of a preexisting drug-resistant subclone. Blood. 2007;110(2):632–9.
50. Arora RS, Arora B. Acute leukemia in children: a review of the current Indian data. South Asian J Cancer. 2016;5(3):155–60.
51. Florento L, Matias R, Tuaño E, Santiago K, Cruz FD, Tuazon A. Comparison of cytotoxic activity of anticancer drugs against various human tumor cell lines using in vitro cell-based approach. Int J Biomed Sci. 2012;8(1):76–80.
52. Anh VC, Kwon JH, Kang JS, Lee HS, Heo CS, Shin JS. New angucycline glycosides from a marine-derived bacterium Streptomyces ardesiacus. Int J Mol Sci. 2022;23(22):13779.
53. Zhou X, Fenical W. The unique chemistry and biology of the piericidins. J Antibiot. 2016;69(8):582–93.
54. Azad SM, Jin Y, Ser HL, Goh BY, Lee LH, Thawai C, et al. Biological insights into the piericidin family of microbial metabolites. J Appl Microbiol. 2022;132(2):772–84.
55. Ooka K, Fukumoto A, Yamanaka T, Shimada K, Ishihara R, Anzai Y, Kato F. Piericidins, novel quorum-sensing inhibitors against Chromobacterium violaceum CV026, from Streptomyces sp. TOHO-Y209 and TOHO-O348. Open J Medi Chem. 2013;3(4):93–9.
56. Kang JE, Han JW, Jeon BJ, Kim BS. Efficacies of quorum sensing inhibitors, piericidin A and glucopiericidin A, produced by Streptomyces xanthocidicus KPP01532 for the control of potato soft rot caused by Erwinia carotovora subsp. Atroseptica. Microbiol Res. 2016;184:32–41.
57. Morgan JM, Duncan MC, Johnson KS, Diepold A, Lam H, Dupzyk AJ, et al. Piericidin A1 blocks Yersinia Ysc Type III secretion system needle assembly. mSphere. 2017;2(1):e00030–17.
58. Li K, Su Z, Gao Y, Lin X, Pang X, Yang B, et al. Cytotoxic minor piericidin derivatives from the actinomycete strain Streptomyces psammoticus SCSIO NS126. Mar Drugs. 2021;19(8):428.
59. Schulze CJ, Bray WM, Loganzo F, Lam MH, Szal T, Villalobos A, et al. Borrelidin B: isolation, biological activity, and implications for nitrile biosynthesis, J Nat Prod. 2014;77(11):2570–4.
60. Kim J, Shin D, Kim SH, Park W, Shin Y, Kim WK, et al. Borrelidins C–E: new antibacterial macrolides from a saltern-derived halophilic Nocardiopsis sp. Mar Drugs. 2017;15(6):166.
61. Lukarska M, Palencia A. Aminoacyl-tRNA synthetases as drug targets. Enzyems. 2020;48:321–50.
62. Fang P, Yu X, Jeong SJ, Mirando A, Chen K, Chen X, et al. Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase. Nat Commun. 2015;31(6):6402.
63. Habibi D, Ogloff N, Jalili RB, Yost A, Weng AP, Ghahary A, et al. Borrelidin, a small molecule nitrile-containing macrolide inhibitor of threonyl-tRNA synthetase, is a potent inducer of apoptosis in acute lymphoblastic leukemia. Invest New Drugs. 2012;30(4):1361–70.
64. Yang Z, He J, Wei X, Ju J, Ma J. Exploration and genome mining of natural products from marine Streptomyces. Appl Microbiol Biotechnol. 2020;104(1):67–76.