INTRODUCTION
Geldanamycin (GDM) is a benzoquinone ansamycin antibiotic. It exhibited anticancer activity by deactivating protein kinase functions in various cancers (Whitesell et al., 1994). The target of GDM is heat shock protein (Hsp90), which induces the proteasomal degradation of target proteins pertinent to cancer (Mimnaugh et al., 1996). However, the utilization of GDM has been limited by its low solubility in water, hepatotoxicity, metabolic instability, and nephrotoxicity (Supko et al., 1995). Therefore, GDM derivatives with better pharmacokinetic profiles have been established (Le Brazidec et al., 2004). Recently, tryptamine-geldanamycin hybrids have been synthesized. These compounds showed anticancer activity against various cancer cell lines and increasing water solubility (Taechowisan et al., 2020). According to these effects, amine-geldanamycin hybrids have been invented. The C17 methoxyl of the GDM molecule can permit the introduction of nucleophilic molecules. Thus, GDM has been a precursor for novel derivatives (Lin et al., 2015; Modi et al., 2011; Supko et al., 1995; Tian et al., 2004; Wrona et al., 2010).
In this study, novel 17-substituted GDM derivatives with (S)-2-amino-3-(1H-indol-3-yl)propan-1-ol, (S)-2-amino-3-phenylpropan-1-ol, and (S)-4-(2-amino-3-hydroxypropyl)phenol were synthesized. Molecular docking was performed to determine the hydrogen-bonding interaction, the binding energy, and the orientation of inhibitors bound to the active site of Hsp90. The absorption, distribution, metabolism, excretion, and toxicity (ADMET) of the compounds was predicted via in silico methods. Their anticancer activity was then evaluated on three cancer cell lines, MDA-MB-231, human cervical carcinoma cells (HeLa), and human hepatocellular carcinoma cells (HepG2) cells, using an (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) (MTT) assay. The water solubility was also carried out.
MATERIALS AND METHODS
The cultivation, extraction of secondary metabolites, including chromatographic fractionation, purification, and structure elucidation of GDM were carried out, as described previously (Taechowisan et al., 2019).
(L)-Tryptophan, (L)-phenylalanine, and (L)-tyrosine were products of Sigma-Aldrich. The 1H and 13C NMR spectra were recorded with a Bruker Avance 300 spectrometer (Bruker, MA). Mass spectra were determined with a micrOTOF (Bruker Daltonics). Melting points were measured by a Stuart Scientific SMP2 melting point apparatus and were uncorrected. The reaction was monitored by TLC, performed on aluminum sheets precoated with silica gel 60 (Darmstadt, Germany). Column chromatography was performed using Merck Kieselgel 60.
General procedure for esterification of amino acids
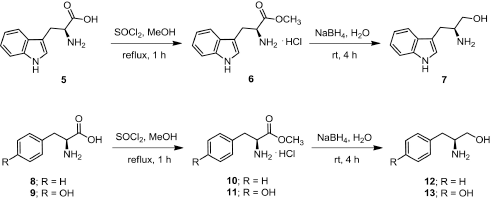
Freshly distilled thionyl chloride (5.5 mmol) was added slowly to a solution of amino acid (5, 8, or 9) (5 mmol) in dry methanol (10 ml) at 0oC and refluxed for 1 hour. The mixture was cooled to room temperature and the solvent was concentrated under vacuum and recrystallized from methanol to provide amino acid methyl ester hydrochloride as white crystals (Fig. 1).
(S)-Methyl 2-amino-3-(1H-indol-3-yl)propanoate hydrochloride (6)
(1.27 g, 99.80%); m.p. 218oC–219oC; 1H-NMR (D2O, 300 MHz) δ 3.34 (dd, J = 4.4, 6.5 Hz, 2H, CH2), 3.68 (s, 3H, CH3), 4.32 (dd, J = 5.9, 6.6 Hz, 1H, CH), 7.07 (td, J = 0.9, 6.9 Hz, 1H, CH), 7.16 (m, 1H, CH), 7.18 (s, 1H, CH), 7.41 (d, J = 8.19 Hz, 1H, CH), 7.48 (d, J = 7.9 Hz, 1H, CH); 13C-NMR (D2O, 75 MHz) δ 25.6, 53.2, 53.5, 105.9, 112.0, 118.0, 119.5, 122.2, 125.3, 126.3, 136.2, 170.4.
(S)-Methyl 2-amino-3-phenylpropanoate hydrochloride (10)
(1.08 g, 100%); m.p. 165oC–166oC; 1H-NMR (D2O, 300 MHz) δ 3.23 (dd, J = 7.4, 14.5 Hz, 1H, CH), 3.35 (dd, J = 5.6, 14.5 Hz, 1H, CH), 3.84 (s, 3H, CH3), 4.43 (dd, J = 5.9, 7.1 Hz, 1H, CH), 7.28–7.45 (m, 5H, ArH); 13C-NMR (D2O, 75 MHz) δ 35.5, 53.5, 54.0, 128.0, 129.2, 129.3, 133.6, 170.0.
(S)-Methyl 2-amino-3-(4-hydroxyphenyl)propanoate hydrochloride (11)
(1.15 g, 99.8%); m.p. 195oC–197oC; 1H-NMR (D2O, 300 MHz) δ 3.16 (dd, J = 7.5, 14.7 Hz, 1H, CH), 3.26 (dd, J = 5.8, 14.7 Hz, 1H, CH), 3.83 (s, 3H, CH3), 4.25 (dd, J = 5.9, 7.3 Hz, 1H, CH), 6.89 (d, J = 8.6 Hz, 2H, CH), 7.16 (d, J = 8.6 Hz, 2H, CH); 13C-NMR (D2O, 75 MHz) δ 34.7, 53.5, 54.1, 115.9, 125.3, 130.8, 155.1, 170.0.
General procedure for reduction of amino acid methyl esters
A solution of amino acid methyl ester (6, 10, or 11) (1 mmol) in THF : H2O (10 : 1) (3 ml) was added to a solution of NaBH4 (4 mmol) in H2O (5 ml) under 0oC. The reaction mixture was stirred at room temperature for 4 hours upon monitoring the process by TLC (Fig. 1). The resulting mixture was acidified with 2 N HCl and extracted with ether. The aqueous phase was adjusted to base with 2 N NaOH and extracted with EtOAc. The organic phases were combined and washed with H2O and then dried over with anh. Na2SO4. The organic phase was concentrated under reduced pressure to provide amino alcohol which was used in the following step without further purification.
(S)-2-Amino-3-(1H-indol-3-yl)propan-1-ol (7)
Light brown oil; (0.12 g); 1H-NMR (CD3OD, 300 MHz) δ 2.71 (dd, J = 7.5, 14.3 Hz, 1H, CH), 2.92 (dd, J = 5.9, 14.3 Hz, 1H, CH), 3.11–3.20 (m, 1H, CH), 3.42 (dd, J = 7.0, 10.8 Hz, 1H, CH), 3.60 (dd, J = 4.8, 10.8 Hz, 1H, CH), 7.02 (td, J = 1.1, 7.0 Hz, 1H, CH), 7.07 (s, 1H, CH), 7.11 (td, J = 1.1, 7.0 Hz, 1H, CH), 7.37 (d, J = 8.0 Hz, 1H, CH), 7.58 (d, J = 7.7 Hz, 1H); 13C-NMR (CD3OD, 75 MHz) δ 30.0, 54.4, 67.0, 112.3, 112.4, 119.52, 119.8, 122.5, 124.4, 128.9, 138.2.
| Figure 1. Scheme of the synthesis of amino alcohol 7, 12, and 13. |
(S)-2-Amino-3-phenylpropan-1-ol (12)
Pale yellow oil; (0.11 g); 1H-NMR (CD3OD, 300 MHz) δ 2.59 (dd, J = 7.7, 13.4 Hz, 1H, CH), 2.79 (dd, J = 6.2, 13.4 Hz, 1H, CH), 3.04–3.13 (m, 1H, CH), 3.41 (dd, J = 6.8, 10.9 Hz, 1H, CH), 3.57 (dd, J = 4.3, 10.9 Hz, 1H, CH), 7.18–7.32 (m, 5H, ArH); 13C-NMR (CD3OD, 75 MHz) δ 40.5, 55.4, 66.2, 127.5, 129.6, 130.3, 139.8.
(S)-4-(2-Amino-3-hydroxypropyl)phenol (13)
Pale orange color oil; (0.14 g); 1H-NMR (CD3OD, 300 MHz) δ 2.50 (dd, J = 7.7, 13.6 Hz, 1H, CH), 2.70 (dd, J = 6.3, 13.6 Hz, 1H, CH), 2.99–3.04 (m, 1H, CH), 3.38 (dd, J = 6.9, 10.8 Hz, 1H, CH), 3.55 (dd, J = 4.4, 10.9 Hz, 1H, CH), 6.76 (d, J = 8.5 Hz, 2H, CH) , 7.05 (d, J = 8.5 Hz, 2H, CH); 13C-NMR (CD3OD, 75 MHz) δ 39.5, 55.6, 66.3, 116.5, 130.4, 131.3, 157.2.
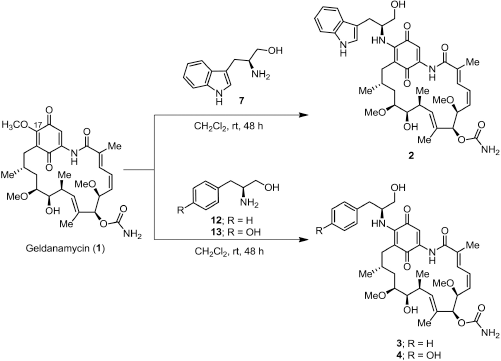
Synthesis of 17-demethoxygeldanamycin derivatives
A solution of amino alcohol (7, 12, or 13) (0.30 mmol) in MeOH (2 ml) was added to a solution of GDM (1) (0.14 mmol) in CH2Cl2 (10 ml). The reaction was kept away from light and stirred at room temperature for 48 hours (Fig. 2). The solvent was removed and diluted with EtOAc and then washed with 1 N HCl, H2O, and brine. The organic phases were combined and dried over with anh. Na2SO4. The organic phase was concentrated under reduced pressure. The crude product was purified by flash column chromatography on silica gel eluting with 40% CH2Cl2 in EtOAc to afford the product as a dark purple solid.
17-((S)-2-Amino-3-(1H-indol-3-yl)propan-1-ol)-17-demethoxygeldanamycin (2)
(0.032 g, 32.1%); m.p. 144oC–146oC; 1H-NMR (CDCl3, 300 MHz) δ 0.86 (d, J = 6.3 Hz, 3H, CH3), 0.97 (d, J = 6.9 Hz, 3H, CH3), 1.74 (m, 4H, hydrocarbon), 1.79 (s, 3H, CH3), 1.97 (s, 3H, CH3), 2.24 (m, 1H, CH), 2.60 (d, J = 13.9 Hz, 1H, CH), 2.73 (m, 1H, CH), 3.16, (d, J = 6.0 Hz, 2H, CH2), 3.25 (s, 3H, CH3), 3.33 (m, 4H, CH, CH3), 3.42 (m, 1H, CH), 3.56 (d, J = 7.5 Hz, 1H, CH), 3.64 (d, J = 4.4 Hz, 2H, CH2), 4.28 (d, J = 9.8 Hz, 1H, CH), 4.52 (m, 1H, CH), 4.93 (br, 2H, NH2), 5.15 (s, 1H, CH), 5.85 (m, 2H, CH), 6.49 (m, 2H, CH), 6.88 (d, J = 11.5 Hz, 1H, CH), 7.17 (m, 4H, CH), 7.37 (d, J = 7.9 Hz, 1H, CH), 7.66 (d, J = 7.7 Hz, 1H, CH), 8.52 (br, 1H, NH), 9.08 (br, 1H, NH); 13C-NMR (CDCl3, 75 MHz) δ 12.3, 12.5, 12.7, 22.8, 27.3, 28.6, 32.2, 34.5, 34.9, 55.4, 56.7, 57.1, 63.1, 72.6, 81.2, 81.3, 81.8, 109.0, 110.2, 111.3, 118.6, 119.7, 119.9, 122.3, 123.5, 126.6, 126.8, 127.6, 132.8, 133.7, 135.0, 135.7, 136.3, 140.8, 145.3, 156.2, 168.3, 180.2, 184.2; HRMS calculated for C39H50N4O9 (M+Na)+ 741.3470, found 741.3473.
17-((S)-2-Amino-3-phenylpropan-1-ol)-17-demethoxygeldanamycin (3)
(0.016 g, 40.6%); m.p. 129oC–131oC; 1H-NMR (CDCl3, 300 MHz) δ 0.84 (d, J = 6.4 Hz, 3H, CH3), 0.98 (d, J = 6.4 Hz, 3H, CH3), 1.75 (m, 4H, hydrocarbon), 1.78 (s, 3H, CH3), 2.02 (s, 3H, CH3), 2.21 (m, 1H, CH), 2.63 (d, J = 13.1 Hz, 1H, CH), 2.73 (m, 1H, CH), 2.93 (dd, J = 6.8, 13.6 Hz, 1H, CH), 3.08 (dd, J = 6.6, 13.6 Hz, 1H, CH), 3.26 (s, 3H, CH3), 3.35 (s, 3H, CH3), 3.42 (m, 1H, CH), 3.55 (dd, J = 1.9, 8.8 Hz, 1H, CH), 3.64 (d, J = 3.8 Hz, 2H, CH2), 4.29 (d, J = 9.8 Hz, 1H, CH), 4.40 (m, 1H, CH), 4.95 (br, 2H, NH2), 5.16 (s, 1H, CH), 5.86 (m, 2H, CH), 6.37 (d, J = 9.1 Hz, 1H, CH), 6.57 (t, J = 11.6 Hz, 1H, CH), 6.92 (d, J = 11.5 Hz, 1H, CH), 7.22 (s, 1H, CH), 7.28 (m, 5H, ArH), 9.10 (br, 1H, NH); 13C-NMR (CDCl3, 75 MHz) δ 12.4, 12.5, 12.8, 23.0, 28.7, 32.2, 34.5, 35.0, 38.2, 56.5, 56.7, 57.1, 63.0, 72.6, 81.1, 81.3, 81.7, 109.0, 109.3, 126.5, 126.9, 128.7, 129.4, 132.8, 133.6, 134.9, 135.8, 136.7, 140.9, 144.9, 156.2, 168.3, 180.4, 184.1; HRMS calculated for C37H49N3O9 (M+Na)+ 702.3361, found 702.3360.
| Figure 2. Scheme of the synthesis of 17-demethoxygeldanamycin derivatives; 17-((s)-2-amino-3-(1H-indol-3-ylpropan-1-ol)-17-demethoxygeldanamycin (2), 17-((s)-2-amino-3-phenylpropan-1-ol)-17-demethoxygeldanamycin (3), and 17-((s)-4-(2-mino-3-hydroxypropyl)phenol)-17-demethoxygeldanamycin (4). |
17-((S)-4-(2-Amino-3-hydroxypropyl)phenol)-17-demethoxygeldanamycin (4)
(0.028 g, 29.2%); m.p. 125oC–127oC; 1H-NMR (CDCl3, 300 MHz) δ 0.89 (d, J = 6.2 Hz, 3H, CH3), 1.01 (d, J = 6.2 Hz, 3H, CH3), 1.78 (m, 2H, CH2), 1.80 (s, 3H, CH3), 2.02 (s, 3H, CH3), 2.14 (m, 2H, CH2), 2.65 (d, J = 13.4 Hz, 1H, CH), 2.75 (m, 1H, CH), 2.94 (d, J = 5.0 Hz, 2H, CH2), 3.27 (s, 3H, CH3), 3.36 (s, 3H, CH3), 3.45 (m, 1H, CH), 3.59 (m, 3H, CH, CH2), 4.30 (d, J = 9.8 Hz, 1H, CH), 4.38 (m, 2H, CH2), 4.79 (br, 2H, NH2), 5.17 (s, 1H, CH), 5.54, (br, 1H, NH), 5.86 (dd, J = 10.6, 10.8 Hz, 2H, CH), 6.37 (d, J = 8.8 Hz, 1H, CH), 6.58 (t, J = 11.3 Hz, 1H, CH), 6.81 (d, J = 8.5 Hz, 2H, CH), 6.93 (d, J = 11.6 Hz, 1H, CH), 7.11 (d, J = 8.5 Hz, 2H, CH), 7.24 (s, 1H, CH), 9.11 (br, 1H, NH); 13C-NMR (CDCl3, 75 MHz) δ 12.5, 12.7, 12.9, 23.1, 28.8, 32.0, 32.4, 34.5, 35.1, 37.1, 56.4, 56.8, 57.3, 62.7, 72.8, 81.3, 81.4, 82.0, 109.1, 109.3, 115.8, 126.8, 127.0, 128.0, 130.8, 133.0, 133.6, 135.1, 135.9, 141.0, 155.2, 156.4, 168.5, 180.5, 184.2; HRMS calculated for C37H49N3O10 (M+Na)+ 718.3310, found 718.3309.
The water solubility of the novel amine-geldanamycin hybrids was determined by comparison with GDM as described previously (Taechowisan et al., 2020).
Molecular docking studies
The two-dimensional structures of GDM and its derivatives were drawn and converted to a SMILES string by ChemDraw software (http://cambridgesoft.com) and the SMILES Translator and Structure Generator (https://cactus.nci.nih.gov/translate/), respectively. The energies of these compounds were optimized and converted to #D format, saved as Protein Data Bank (PDB) files using UCSF Chimera v.1.14 (University of California, Oakland, CA), and further used for docking studies.
The three-dimensional structure of Hsp90 with the cocrystallized GDM (PDB ID: 1OSF) was retrieved from the Research Collaborator for Structural Bioinformatics PDB and chosen for molecular docking studies. The crystal structure of 1OSF was employed for docking GDM and amine-geldanamycin hybrids to obtain reliable predictions of ligand bindings. The water molecules were removed from the crystal structure using Discovery Studio software followed by the addition of Gasteiger charges to targets.
Docking simulations of Hsp90 target (1OSF) with 17-DMAG, GDM, and amine-geldanamycin hybrids were undertaken using AutoDock Vina for predicting the binding site of Hsp90 and its ligands. The target conformation was set as a rigid unit, while the ligands were conceded to be flexible and adoptable to the target. Vina sought the lowest binding affinity conformations and provided five different conformations for the Hsp90 target. The lowest-binding-energy docking poses of each compound were selected. AutoDock Vina was processed using an exhaustiveness of four and a grid box with center dimensions of x = 30.2535, y = 45.3258, and z = 52.7852 with a size of X = 41.3526, Y = 43.2578, and Z = 50.8467 for 1OSF. The UCSF Chimera v.1.14 was chosen for visual inspection and preparations. Protein–ligand interactions were analyzed with the aid of Chimera and LigPlot v.4.5.3.
Prediction of ADMET by computational analysis
The computational prediction of the compounds was performed using the online software SwissADME (http://swissadme.ch) and Pre-ADMET (https://preadmet.bmdrc.kr) to obtain relative results for pharmacokinetic profile (ADMET) of the molecules. The ADMET profiles of the novel amine-geldanamycin hybrids were analyzed in comparison with that of GDM.
Cytotoxicity assay
Two normal cell lines [African green monkey kidney cells (Vero) and rhesus monkey kidney cells (LLC-MK2)] and three cancer cell lines [human breast carcinoma cells (MDA-MB-231); HepG2; and HeLa (from the Korean Cell Line Bank))] were grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% in a 37°C, 5% CO2 humidified incubator. Cytotoxicity assay was carried out as described in a previous publication (Taechowisan et al., 2020).
Statistical analysis
Each result represents the means ± standard deviation of three experiments. SPSS v.16.0 (SPSS Inc., Chicago, IL) software was used for data analysis. Comparisons between the two groups were analyzed using the two-tailed Dunnett t-tests treated compound 1 as a control group. A p-value < 0.05 was considered to indicate statistical significance.
RESULTS
Hsp90, PBD ID: 1OSF, was selected for molecular docking studies. Comparative docking of 1OSF with 17-DMAG and the tested compounds was performed to support the in vivo antiviral activity.
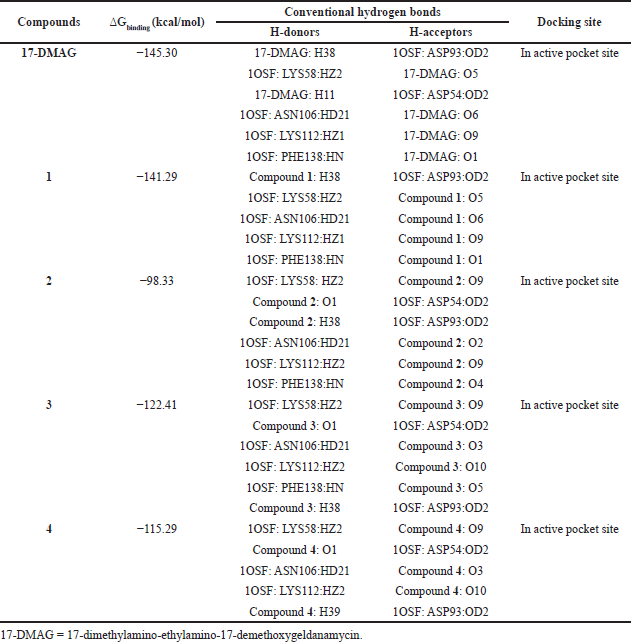
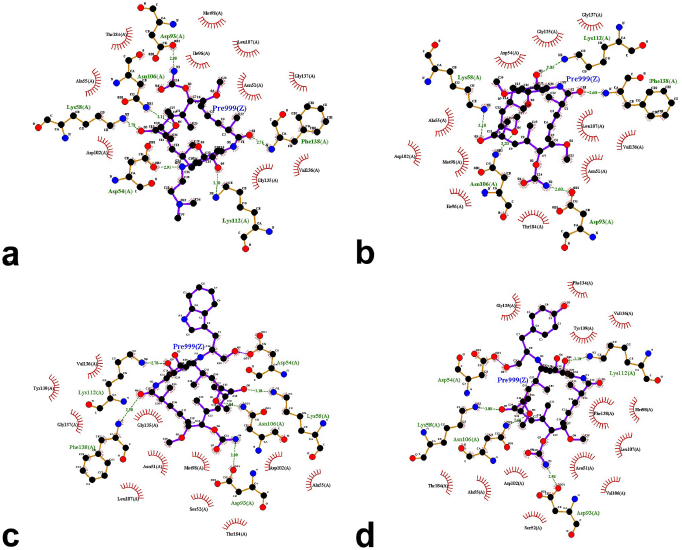
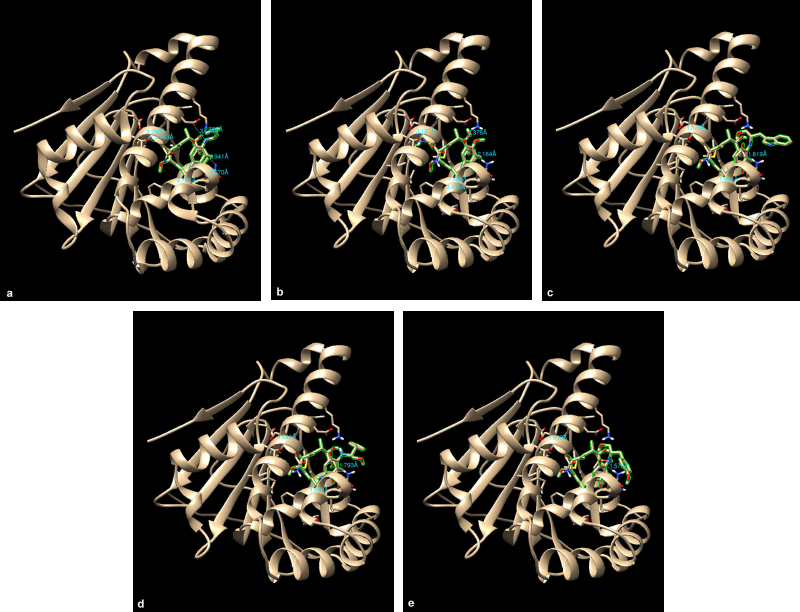
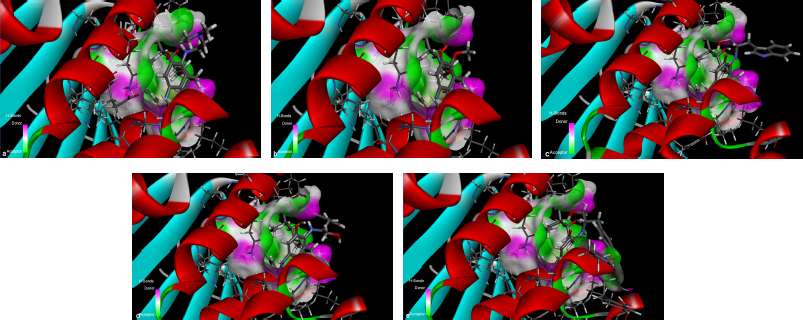
The results from the docking study (as shown in Table 1) revealed that 17-DMAG interacted through six hydrogen bonds with Asp54, Lys58, Asp93, Asn106, Lys112, and Phe138 to the N-terminal domain pocket of Hsp90, with a binding energy of -145.307 kcal/mol. GDM bridged five hydrogen bonds with Lys58, Asp93, Asn106, Lys112, and Phe138 to the N-terminal domain pocket of Hsp90, with a binding energy of −141.296 kcal/mol. Compounds 2 and 3 bridged six hydrogen bonds with Asp54, Lys58, Asp93, Asn106, Lys112, and Phe138 to the N-terminal domain pocket of Hsp90, with a binding energy of −98.33 and −122.41 kcal/mol, respectively. Compound 4 bridged five hydrogen bonds with Asp54, Lys58, Asp93, Asn106, and Lys112 to the N-terminal domain pocket of Hsp90, with a binding energy of −115.29 kcal/mol. The protein–ligand interaction plots generated using LigPlot (Fig. 3) indicated that all ligands exhibited hydrogen bonding similar to that observed by AutoDock Vina. Also, all compounds exhibited similar hydrophobic interactions with the same residues Asn51, Ala55, Met98, Asp102, Leu107, Gly135, Val136, and Thr184, with a few exceptions in the case of compounds 2, 3, and 4, which were due to distinction in the position of compounds within the active site of Hsp90. The docking presentations of crystal structures are displayed in Figure 4. The protein–ligand interactions revealed that the 17-DMAG, GDM, and its derivatives exhibited interactions with five and six hydrogen bonds to the other amino acids in the active site pocket, respectively; however, 17-DMAG and GDM showed good docking energy, −145.30 ,and −141.29 kcal/mol, respectively, while the docking energy of the GDM derivatives was −98.33 to −122.41 kcal/mol, which was lower than those of 17-DMAG and GDM. The molecular docking data provides structural insights about the binding interactions of ligands in the active site of the target protein.
| Table 1. Molecular docking results of 17-DMAG, GDM (1), and its derivatives (2–4) to Hsp90 (1OSF). |
The 17-DMAG, GDM, and its derivatives were completely enfolded in the entire active pocket of Hsp90 (Fig. 5). The topology of the active site of Hsp90 was similar in both 17-DMAG and GDM binding, which was lined with interacting amino acids, as predicted by LigPlot (Fig. 3). However, the orientation of compounds 2, 3, and 4 was perpendicular to the grove of the N-terminal domain of Hsp90 (Figs. 3 and 4). These resulted in lower binding energy than those of 17-DMAG and GDM.
The solubility in water of GDM was found to be 0.152 mM (Table 2), while the solubility in water of compounds 2, 3, and 4 was 5.571 mM, 1.963 mM, and 1.918 mM, respectively, approximately 36.65, 12.91, and 12.62 times, respectively, higher than that of GDM. These data suggest that the conjugation of an amine moiety to GDM at the C17-position notably improved its solubility in water.
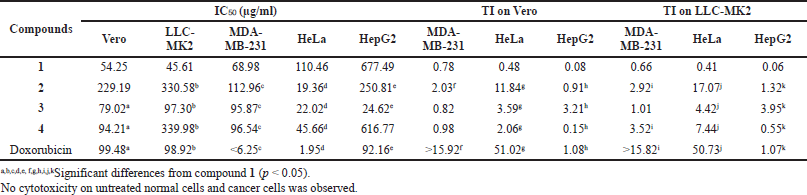
GDM and its derivatives were also assessed for cytotoxicity against two normal cell lines (Vero and LLC-MK2 cells) and three cancer cell lines (MDA-MB-231, HeLa, and HepG2) using the MTT assay. Compound 2 showed weak cytotoxic activity toward Vero and LLC-MK2 cells with IC50 values of 229.19 and 330.58 µg/ml, respectively (Table 3). However, this compound exhibited higher cytotoxicity activity to HeLa cells, with IC50 values of 19.36 µg/ml. In addition, compounds 3 and 4 were also active against HeLa cells, with IC50 values of 22.02 and 45.66 µg/ml, respectively. Among these compounds, only compound 3 was active against HepG2 cells, with IC50 values of 24.62 µg/ml. However, these compounds showed lower cytotoxic activity toward MDA-MB-231 than GDM. The cytotoxicity of these compounds against HeLa cells and MDA-MB231, with IC50 values of doxorubicin (standard), was 1.95 and <6.25 µg/ml, respectively. In addition, no cytotoxicity on untreated normal cells and cancer cells was observed (data not shown). The therapeutic index (TI) of the tested compounds differed from each other. In both normal cell types (Vero and LLC-MK2) tested here, the TI values were in the following order: doxorubicin > compound 2 > compound 3 > compound 4 > compound 1. The TI for doxorubicin was highest in comparison with GDM and its derivatives. Compound 2 showed TI of 11.84 and 17.07 relative to Vero and LLC-MK2, respectively, when compared to HeLa cells, while compound 1 showed the lowest TI among the compounds tested here, with values of 0.08 and 0.06 relative to Vero and LLC-MK2, respectively, when compared to HepG2 cells. It suggested that these amine-geldanamycin hybrids were more effective against some cancer cells than normal cells. Consequently, these compounds can display potential application in anticancer chemotherapy at recommended dosages.
| Figure 3. LigPlot showing hydrogen-bonding interactions (the green dashed lines) and hydrophobic contacts (the red crescents with the bristles) for the ligand: 17-DMAG (a), compound 1 (b), compound 2 (c), and compound 4 (d) molecules in active pocket of Hsp90. |
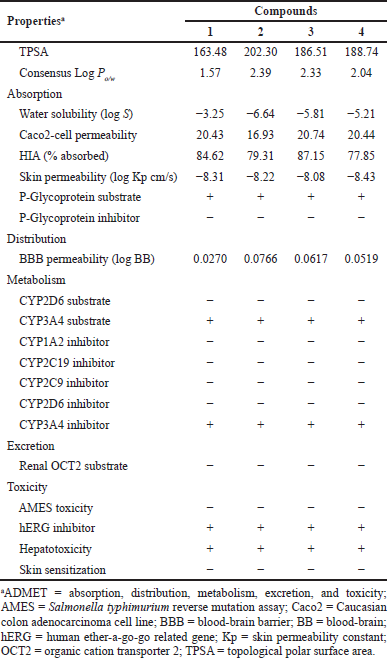
The ADMET parameters of GDM and its derivatives were presented in Table 4. The TPSA of GDM and its derivatives were greater than 140, which suggested that these compounds were high polar molecules. They were predicted as having ideal lipophilicity (average of Log Po/w ≤ 5). This result suggested that these compounds have good absorption and permeation. The absorption level of the compounds was predicted by Caco2-cell permeability, skin permeability, human intestinal absorption (HIA), and P-glycoprotein inhibitor or substrate. When the predicted value of Caco2-cell permeability is ≥ 0.90, the compound is easy to absorb. GDM and its derivatives were predicted to have high Caco2-cell permeability. These compounds were predicted to have high skin permeability (the log Kp < −2.5). The compounds were predicted to be substrates of P-glycoprotein; they may be excreted from cells by P-glycoprotein. They were also predicted not to be P-glycoprotein inhibitors. A compound with an HIA value less than 30% is considered to be poorly absorbed. These compounds were predicted to have good absorption. These compounds were predicted to moderately cross the blood–brain barrier (−1 < logBB < 0.3). Cytochrome P450 subtype is an enzyme involved in drug metabolism in the liver. The results showed that GDM and its derivatives were substrates for CYP3A4 but not for CYP2D6. GDM and its derivatives were also predicted to be CYP3A4 inhibitor. This suggested that GDM and its derivatives may be metabolized in the liver. The results also suggest that GDM and its derivatives may be nonmutagens in the Technique to determine metagenic potential (AMES) test, but they may deactivate the human ether-a-go-go related gene (hERG) channel and may have cardiotoxicity.
Thus, the predicted results suggest that the ADMET profiles of amine-geldanamycin hybrids are similar to those of GDM. However, they were a cytochrome substrate and inhibitor, which may cause hepatotoxicity. In addition, they may be a cause for neurotoxicity and Central Nervous System (CNS) disease.
| Figure 4. Crystal structure superimposed on Hsp90 (1OSF) docked 17-DMAG (a), compound 1 (b), compound 2 (c), compound 3 (d), and compound 4 (e). |
DISCUSSION
GDM was the first benzoquinone ansamycin antibiotic generated by Streptomyces hygroscopicus var. Geldanus, which was discovered in 1970 (Johnson et al., 2010). Recently, it was also isolated from Streptomyces zerumbet W14 (Taechowisan et al., 2019). It was an inhibitor of Hsp90, which bound to the Adenosine Triphosphate (ATP)-binding pocket. For the past decade, Hsp90 has been a candidate target for cancer therapy because it plays an important role in the stabilization of the cancer cell proteins (Chatterjee and Burns, 2017). Although GDM is a potent cell growth inhibitor, it is hampered by severe hepatotoxicity and insolubility in water (Fukuyo et al., 2009; Supko et al., 1995). Accordingly, considerable efforts have gone into altering the structure of this antibiotic in order to reduce hepatotoxicity and increase water solubility. Several derivatives of GDM are in clinical trials, for example, 17-allylamine-17-demethoxygeldanamycin (Banerji et al., 2005; Goetz et al., 2005; Heath et al., 2008; Ramanathan et al., 2005; Solit et al., 2008), dimethylaminoethylamino17-DMAG (Grem et al., 2005), and 17-allylamino-17-demethoxygeldanamycin hydroquinone hydrochloride (Hanson and Vesole, 2009; Sydor et al., 2006). However, the drug-related toxicity of these compounds was unfavorable, as they caused both cardiac and liver toxicity (Lancet et al., 2006; Sequist and Janne, 2007). Meanwhile, novel GDM derivatives have been synthesized and are excellent tools for exploring its biological activities.
In this study, three new GDM derivatives, 17-((S)-2-amino-3-(1H-indol-3-ylpropan-1-ol)-17-demethoxygeldanamycin (2), 17-((S)-2-amino-3-phenylpropan-1-ol)-17-demethoxygeldanamycin (3), and 17-((S)-4-(2-amino-3-hydroxypropyl)phenol)-17-demethoxygeldanamycin (4), were synthesized from GDM (1). According to molecular docking studies, these compounds bound to the ATP pocket of Hsp90. The binding mode of these compounds using molecular docking analysis has predicted the protein–ligand contacts and their interaction strength of these compounds along with 17-DMAG, the control compound in the active site of Hsp90 (1OSF). These GDM derivatives exhibited a similar posture to that of 17-DMAG and GDM in the binding pocket. The binding energy of these GDM derivatives is in the range from −98.33 to −122.41 kcal/mol, in comparison to the binding energy of −145.30 kcal/mol for 17-DMAG and −141.29 kcal/mol for GDM, bound to Hsp90. The interaction plots displayed by LigPlot indicated that the GDM derivatives exhibit hydrogen bonding most similar to that observed in 17-DMAG, and they exhibited highly conserved hydrophobic interactions with the same amino acid residues.
| Figure 5. Orientation of 17-DMAG (a), compound 1 (b), compound 2 (c), compound 3 (d), and compound 4 (e) molecules in active pocket of Hsp90. |
| Table 2. Water solubility of GDM (1) and its derivatives (2–4). |
| Table 3. Cytotoxicity activity (IC50) of GDM (1) and its derivatives (2–4). |
Similar to the findings of previous studies, the molecular docking in the present study has shown that the pocket of Hsp90 is composed of mixture of charged, polar, and hydrophobic amino acids. These residues comprised Asn51, Asp54, Ala55, Lys58, Asp93, Met98, Asn106, Leu107, Lys112, Gly135, Phe138, and Thr184. Therefore, the bottom of the pocket altered increasingly hydrophobic. However, it kept one charged residue (Asp93) and one polar residue (Thr184) at the deepest portion of the pocket. These residues could form hydrophobic interaction with the ligands (Abbasi et al., 2017; Stebbins et al., 1997; Teo et al., 2015). The ADMET parameters of the compounds were carried out by two predictive software. The relationship between the structure and toxicity of these compounds was preliminarily evaluated. These compounds have good absorption and permeations; they are the substrate of P-glycoprotein and are comfortably transported in the body and may cause hepatotoxicity. Several studies reported that the benzoquinone moiety presented in these compounds is inducible for hepatotoxicity, by redox metabolism of cytochrome P450 and glutathione formation, both of which caused by Hsp90 deactivation (Cysyk et al., 2006; Guo et al., 2008). This information anticipates the theoretical bases of toxicity prediction of these compounds.
| Table 4. Predicted ADMET properties of GDM (1) and its derivatives (2–4). |
The toxicity and solubility in water of GDM have been a marked restriction to develop for chemotherapy use. It has incentived researchers to develop less toxic GDM derivatives. In the present study, the experimental basis was carried out; compounds 2, 3, and 4 showed the greater increase in water solubility; especially compound 2, which has the greatest solubility in water, showed less cytotoxicity than GDM in normal cell lines and presented greater toxicity to some cancer cell lines. The likelihood of use of this compound should be increased in future studies.
The introduction of an amine group at the C17-position of GDM did not interfere with the binding to Hsp90, but it greatly decreased the cytotoxicity and increased the water solubility. As indicated by the crystal structure of the GDM-Hsp90 complex (Stebbins et al., 1997), the substitution of the C17 methoxyl of GDM is revealed on the external cavity of the Hsp90 Protei, while differences in the substituents of GDM are critical for the interaction with Hsp90. In the report by Li et al. (2010), 17-amino-17-demethoxygeldanamycin derivatives have a great potential for anticancer activity, while the 19-substituted GDM modification was not a possibility in terms of anticancer agents. The substitution of the group at C19 methoxyl of GDM could obstruct the binding to Hsp90 by the steric hindrances (Li et al. 2010). This result motivates the researchers to inspect that Hsp90 could be a target for anticancer therapy and that GDM and its derivatives have a great potential for anticancer activity by deactivating Hsp90 functions in cancer cells. Doxorubicin is an antibiotic used in the treatment of cancers. In this study, it was used as the positive control. However, doxorubicin exhibited greater cytotoxicity against MDA-MB231 and HeLa cells than GDM and its derivatives. The mechanisms of anticancer activity of doxorubicin were (i) disruption of topoisomerase-II and intercalation of DNA and (ii) generation of free radicals and damage to cellular membranes, proteins, and DNA, which were different from GDM and its derivatives (Gewirtz, 1999). In addition, the restriction of the use of doxorubicin was cardiotoxicity, and doxorubicin resistance was also a problem (Thorn et al., 2011; Weiss, 1992). Therefore, the development of Hsp90 inhibitors may become a universal chemotherapeutic strategy for cancers. This study will help researchers to uncover structural modifications of the compounds to improve their biological activities.
CONCLUSION
Novel amine-geldanamycin hybrids have been synthesized, and they exhibited improved water solubility. These compounds have potent cytotoxic effects on some cancer cells, especially HeLa cells, with low cytotoxic activity against normal cells. These results suggest that these amine-geldanamycin hybrids could be considered a new choice for the treatment of some cancers.
ACKNOWLEDGMENTS
This research project was funded by the Faculty of Science, Silpakorn University, Nakhon Pathom, Thailand.
AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
Abbasi M, Sadeghi-Aliabadi H, Amanlou M. Prediction of new Hsp90 inhibitors based on 3,4-isoxazolediamide scaffold using QSAR study, molecular docking and molecular dynamic simulation. DARU, 2017; 25(1):17. CrossRef
Banerji U, O’Donnell A, Scurr M, Pacey S, Stapleton S, Asad Y, Simmons L, Maloney A, Raynaud F, Campbell M, Walton M, Lakhani S, Kaye S, Workman P, Judson I. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol, 2005; 23(18):4152–61. CrossRef
Chatterjee S, Burns TF. Targeting heat shock proteins in cancer: a promising therapeutic approach. Int J Mol Sci, 2017; 18(9):1978. CrossRef
Cysyk RL, Parker RJ, Barchi JJ Jr, Steeg PS, Hartman NR, Strong JM. Reaction of geldanamycin and C17-substituted analogues with glutathione: product identifications and pharmacological implications. Chem Res Toxicol, 2006; 19(3):376–81. CrossRef
Fukuyo Y, Hunt CR, Horikoshi N. Geldanamycin and its anti-cancer activities. Cancer Lett, 2009; 290(1):24–35. CrossRef
Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem Pharmacol, 1999; 57(7):727–41. CrossRef
Goetz MP, Toft D, Reid J, Ames M, Stensgard B, Safgren S, Adjei AA, Sloan J, Atherton P, Vasile V, Salazaar S, Adjei A, Croghan G, Erlichman C. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J Clin Oncol, 2005; 23(6):1078–87. CrossRef
Grem JL, Morrison G, Guo XD, Agnew E, Takimoto CH, Thomas R, Szabo E, Grochow L, Grollman F, Hamilton JM, Neckers L, Wilson RH. Phase I and pharmacologic study of 17-(allylamino)-17-demethoxygeldanamycin in adult patients with solid tumors. J Clin Oncol, 2005; 23(9):1885–93. CrossRef
Guo W, Reigan P, Siegel D, Ross D. Enzymatic reduction and glutathione conjugation of benzoquinone ansamycin Hsp90 inhibitors: relevance for toxicity and mechanism of action. Drug Metab Dispos, 2008; 36(10):2050–7. CrossRef
Hanson BE, Vesole DH. Retaspimycin hydrochloride (IPI-504): a novel heat shock protein inhibitor as an anticancer agent. Expert Opin Investig Drugs, 2009; 18(9):1375–83. CrossRef
Heath EI, Hillman DW, Vaishampayan U, Sheng S, Sarkar F, Harper F, Gaskins M, Pitot HC, Tan W, Ivy SP, Pili R, Carducci MA, Erlichman C, Liu G. A phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with hormone-refractory metastatic prostate cancer. Clin Cancer Res, 2008; 14(23):7940–6. CrossRef
Johnson VA, Singh EK, Nazarova LA, Alexander LD, McAlpine SR. Macrocyclic inhibitors of hsp90. Curr Top Med Chem, 2010; 10(14):1380–402. CrossRef
Lancet J, Baer MR, Gojo I, Burton M, Quinn M, Tighe SM, Bhalla K, Kersey K, Wells S, Zhong Z. In phase 1, pharmacokinetic (PK) and pharmacodynamic (PD) study of intravenous alvespimycin (KOS-1022) in patients with refractory hematological malignancies; ASH Annual Meeting, San Diego, CA, 2006. CrossRef
Le Brazidec JY, Kamal A, Busch D, Thao L, Zhang L, Timony G, Grecko R, Trent K, Lough R, Salazar T, Khan S, Burrows F, Boehm MF. Synthesis and biological evaluation of a new class of geldanamycin derivatives as potent inhibitors of Hsp90. J Med Chem, 2004; 47(15):3865–73. CrossRef
Li YP, Shan GZ, Peng ZG, Zhu JH, Meng S, Zhang T, Gao LY, Tao PZ, Gao RM, Li YH, Jiang JD, Li ZR. Synthesis and biological evaluation of heat-shock protein 90 inhibitors: geldanamycin derivatives with broad antiviral activities. Antivir Chem Chemother, 2010; 20:259–68. CrossRef
Lin Z, Peng R, Li Z, Wang Y, Lu C, Shen Y, Wang J, Shi G. 17-ABAG, a novel geldanamycin derivative, inhibits LNCaP-cell proliferation through heat shock protein 90 Inhibition. Int J Mol Med, 2015; 36(2):424–32. CrossRef
Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem, 1996; 271(37):22796–801. CrossRef
Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, D’Andrea G, Dickler M, Moynahan ME, Sugarman S, Ma W, Patil S, Norton L, Hannah AL, Hudis C. HSP90 inhibition is effective in breast cancer: a phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin Cancer Res, 2011; 17(15):5132–9. CrossRef
Ramanathan RK, Trump DL, Eiseman JL, Belani CP, Agarwala SS, Zuhowski EG, Lan J, Potter DM, Ivy SP, Ramalingam S, Brufsky AM, Wong MK, Tutchko S, Egorin MJ. Phase I pharmacokinetic-pharmacodynamic study of 17-(allylamino)-17-demethoxygeldanamycin (17AAG, NSC 330507), a novel inhibitor of heat shock protein 90, in patients with refractory advanced cancers. Clin Cancer Res, 2005; 11(9):3385–91. CrossRef
Sequist L, Janne P, Sweeney J. In inhibitor IPI-504, in patients with relapsed and/or refractory stage IIIb or stage IV non-small cell lung cancer (NSCLC) stratified by EGFR mutation status. AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics, Philadelphia, PA, 2007.
Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, Teitcher J, Wolchok JD, Germino FJ, Krown SE, Coit D, Rosen N, Chapman PB. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res, 2008; 14(24):8302–7. CrossRef
Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP. Crystal structure of an Hsp90-geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell, 1997; 89(2): 239–50. CrossRef
Supko JG, Hickman RL, Grever MR, Malspeis L. Preclinical pharmacologic evaluation of geldanamycin as an antitumor agent. Cancer Chemother Pharmacol, 1995; 36(4):305–15. CrossRef
Sydor JR, Normant E, Pien CS, Porter JR, Ge J, Grenier L, Pak RH, Ali JA, Dembski MS, Hudak J, Patterson J, Penders C, Pink M, Read MA, Sang J, Woodward C, Zhang Y, Grayzel DS, Wright J, Barrett JA, Palombella VJ, Adams J, Tong JK. Development of 17-allylamino-17-demethoxygeldanamycin hydroquinone hydrochloride (IPI-504), an anti-cancer agent directed against Hsp90. Proc Natl Acad Sci USA, 2006; 103(46):17408–13. CrossRef
Taechowisan T, Puckdee W, Phutdhawong WS. Streptomyces zerumbet, a novel species from Zingiber zerumbet (L.) smith and isolation of its bioactive compounds. Adv Microbiol, 2019; 9(3):194–219. CrossRef
Taechowisan T, Samsawat T, Puckdee W, Phutdhawong WS. Cytotoxicity activity of geldanamycin derivatives against various cancer cell lines. J Appl Pharm Sci, 2020; 10(06):12–21. CrossRef
Teo RD, Dong SS, Gross Z, Gray HB, Goddard WA. Computational predictions of corroles as a class of Hsp90 inhibitors. Mol Biosyst, 2015; 11(11):2907–14. CrossRef
Thorn CF, Oshiro C, Marsh S, Hernandez-Boussard T, McLeod H, Klein TE, Altman RB. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet Genomics, 2011; 21(7):440–6. CrossRef
Tian ZQ, Liu Y, Zhang D, Wang Z, Dong SD, Carreras CW, Zhou Y, Rastelli G, Santi DV, Myles DC. Synthesis and biological activities of novel 17-aminogeldanamycin derivatives. Bioorg Med Chem, 2004; 12(20):5317–29. CrossRef
Weiss RB. The anthracyclines: will we ever find a better doxorubicin? Semin Oncol, 1992; 19(6):670–86.
Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A, 1994; 91(18):8324–8. CrossRef
Wrona IE, Gozman A, Taldone T, Chiosis, G, Panek JS. Synthesis of reblastatin, autolytimycin, and non-benzoquinone analogues: potent inhibitors of heat shock protein 90. J Org Chem, 2010; 75(9):2820–35. CrossRef