INTRODUCTION
Lipases (Triacylglycerol acylhydrolase, EC 3.1.1.3) are water soluble enzymes, which catalyze a wide range of reactions, including hydrolysis, interesterification, alcoholysis, acidolysis, esterification, and aminolysis. These lipases could hydrolyze triacylglycerols to release free fatty acids and glycerol (Javed et al., 2018; Priji et al., 2015). Lipases are ubiquitous in nature and widely distributed in plants, animals, and microorganisms, such as bacteria, yeasts, and fungi. The bacterial lipases are more economical and stable (Andualema and Gessesse, 2012; Ghasemi et al., 2010). Thus, the bacterial lipases are used extensively in food and dairy industry for the hydrolysis of milk fat, cheese ripening, flavor enhancement, and lipolysis of butter fat and cream. They are also used in textile industry to increase fabric absorbency (Andualema and Gessesse, 2012), for the synthesis of biodegradable polymers or compounds, in the energy industry for the synthesis of biodiesel, in the detergent industry as additive in washing powder, and different transesterification reactions (Javed et al., 2018; Liu et al., 2011). Many microorganisms in the genera Bacillus, Burkholderia, Corynebacterium, Geobacillus, Idiomarina, Oceanobacillus, Pseudomonas, Staphylococcus, and Virgibacillus have been reported to produce lipase (Abol-Fotouh et al., 2016; Daroonpunt et al., 2016a; Dey et al., 2014; Gayathri et al., 2013; Li et al., 2014; Phoottosavako et al., 2015; Yang et al., 2016; Yele and Desai, 2015). Bacillus subtilis K-C3 was reported that it could be used as a starter culture to enhance the fermentation of shrimp paste (Pongsetkul et al., 2018). The quality of shrimp paste associated with the formation of free fatty acids in lipolysis, which develops of the flavor or aroma (Itou et al., 2006).
The traditional fermented shrimp paste (Ka-pi), a purple-pink to dark brown color, strong odor and paste-like consistency is widely consumed as seasoning ingredient in Thailand. In Southeast Asia, there are related fermented fish products such as bagoong (Philippines), shiokara (Japan), mam ruoc (Vietnam), terasi (Indonesia), ngapi (Myanmar), and belacan (Malaysia) (Chaijan and Panpipat, 2012; Wittanalai et al., 2011). Many microorganisms are found in shrimp paste and their extracellular lipases play a major role in shrimp paste products (Camacho et al., 2009). The aim of this research was to isolate, identify, and screen the lipolytic activity of bacteria from Thai shrimp paste.
MATERIALS AND METHODS
Sources and isolation methods
Five shrimp paste samples collected from Mahachai market, Samut Sakhon province, Thailand, were used for the isolation. Bacterial strains were isolated by spread plate technique using 1 g of the shrimp paste sample diluted in 99-ml nutrient broth (NB) supplemented with 5% NaCl (w/v). This was then 10-fold serially diluted with modified NB solution and 0.1 ml of each dilution was spread on a modified nutrient agar (NA) plate and incubated at 37°C for 48–72 hours. Colonies which showed a different appearance were picked up and then were transferred to nutrient slant.
Primary screening of lipolytic activity
All of the isolated bacteria were screened on lipolytic agar (Barrow and Feltham, 1993) composed of 1% (w/v) peptone, 0.5% (w/v) yeast extract, 0.01% (w/v) CaCl2.2H2O, 2% (w/v) agar and 1% (v/v) of tributyrin, Tween 20, Tween 40, Tween 60, or Tween 80 and incubated at 37°C for 3–5 days. Isolated halophilic bacteria colonies that showed an opaque zone around the colony (potentially positive for lipase activity) were selected for further study.
Identification methods
Phenotypic characterization
The morphological and cultural characteristics were determined as previously described (Barrow and Feltham, 1993; Daroonpunt et al., 2016b; Leifson, 1963). The isolates were cultivated on NA plates at 37°C for 2–3 days and then examined for the colony and cell characteristics, such as the color and shape and Gram staining of the cells. Acid production from carbon sources was evaluated in basal medium as described by Gordon et al. (1973). The hydrolysis of gelatin, starch, and arginine by each isolate was determined (Barrow and Feltham, 1993) and its ability to grow in different salinity [0%, 1%, 3%, 5%, 7%, and 10% (w/v) NaCl], pH (5–10, interval of 0.5) and temperature (20°C, 25°C, 30°C, 40°C, 45°C, and 50°C) levels was also observed.
Genotypic characterization
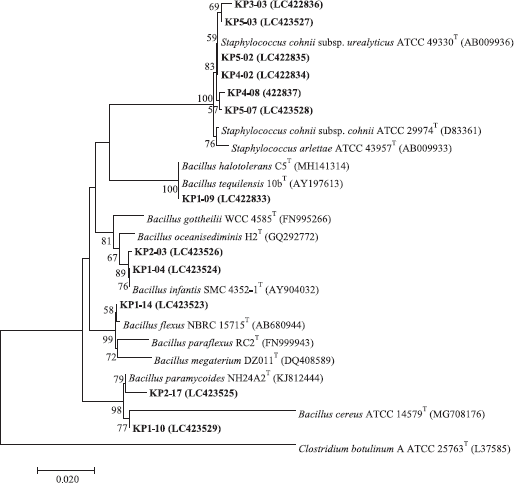
The 16S rRNA gene fragment was amplified by polymerase chain reaction (PCR) as previously described (Yamada et al., 2000). The PCR products were resolved and checked by gel electrophoresis in comparison with a 1-kb DNA marker. The PCR products were sent to Macrogen, Korea, for commercial sequencing using the 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGAC TT-3′) primers. The obtained sequences were checked for homology to known sequences using the standard BLASTn sequence similarity searching program from the EzBiocloud server (Yoon et al., 2017). Multiple alignments of the obtained sequences were performed with the BioEdit program. The neighbor-joining tree (Saitou and Nei, 1987) was then constructed using the MEGA 7 programs (Kumar et al., 2016) and the confidence value of branches of the phylogenetic tree was determined using the bootstrap analysis (Felsenstein, 1985) based on 1,000 replications.
Assay for lipolytic activity
Lipase activity was assayed by measuring the micromole of p-nitrophenol released from p-nitrophenyl butyrate (p-NPB) or p-nitrophenyl palmitate (p-NPP) as the modified colorimetric method (Lee et al., 1999). The substrates of p-NPB or p-NPP were dissolved in ethanol and mixed with 900 μl of universal buffer (Britton–Robinson buffer, pH 8.0) to give a final concentration of 1 mM. After pre-incubation for 5 minutes, the reaction was initiated by adding 50 μl of enzyme solution to 50 μl of substrate solution, and incubated at 37°C for 30 minutes. A 100 μl of 20% sodium dodecyl sulfate was added to stop the reaction and the amount of p-nitrophenol (p-NP) released was measured at 410 nm against a blank. One unit (U) was defined as the amount of enzyme liberating 1 mmol of p-NP per minute under the standard assay conditions.
RESULTS AND DISCUSSION
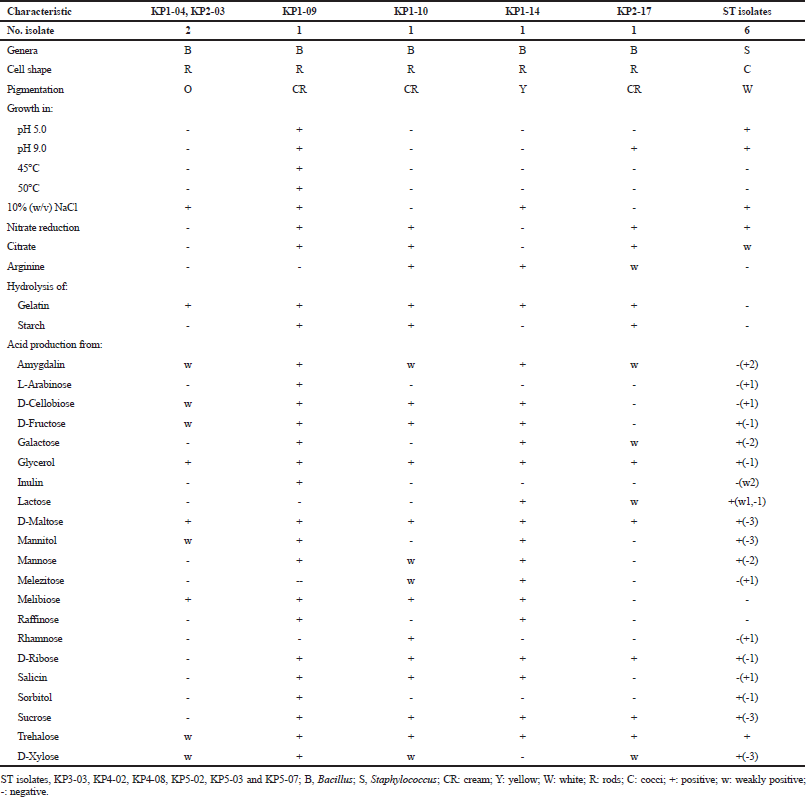
Twelve bacterial strains that produced extracellular lipase were isolated from five samples of shrimp paste collected from Mahachai market, Samut Sakhon province, Thailand. These 12 isolates (KP1-04, KP1-09, KP1-10, KP1-14, KP2-03, KP2-17, KP3-03, KP4-02, KP4-08, KP5-02, KP5-03, and KP5-07) showed opaque halos of calcium oleate on lipolytic agar when Tween was used (Garcia-Lepe et al., 1997), while one strain KP4-08 showed positive for the hydrolysis of tributyrin. All of them were Gram-positive bacteria, and the six rod-shaped isolates were belonged to the genus Bacillus and the six coccal isolates were belonged to the genus Staphylococcus based on their phenotypic characteristics and 16S rRNA gene sequence analysis (Table 1). They grew at 20°C and in 5% NaCl. All produced acid from D-glucose. Their differential phenotypic characteristics are presented in Table 1.
Bacillus isolates, KP1-04 and KP2-03, showed irregular and orange colonies and grew in 0%–10% (w/v) NaCl, pH 6.0–8.0, and at 20°C–40°C. They showed positive for gelatin hydrolysis but negative for nitrate reduction, arginine, citrate utilization, and starch hydrolysis. Acid is produced from amygdalin, cellobiose, D-fructose, glycerol, D-maltose, mannitol, melibiose, trehalose, and xylose but not from L-arabinose, galactose, inulin, lactose, mannose, melezitose, raffinose, rhamnose, D-ribose, salicin, sorbitol, and sucrose (Table 1). Based on the 16S rRNA gene sequence (1,399 and 1,386 bp, respectively), they were identified as B. infantis (Fig. 1) from their 99.64% and 99.57% sequence similarity to B. infantis SMC 4352-1T (Ko et al., 2006). Isolate KP1-09 showed irregular and cream colonies and grew in 0%–10% (w/v) NaCl, pH 5.0–9.0, and at 20°C–50°C. It showed positive for hydrolysis of gelatin and starch, citrate utilization, nitrate reduction but negative for arginine hydrolysis. Acid is produced from amygdalin, L-arabinose, cellobiose, D-fructose, galactose, glycerol, inulin, D-maltose, mannitol, mannose, melibiose, raffinose, D-ribose, salicin, sorbitol, sucrose, trehalose, and D-xylose but negative for lactose, melezitose, and rhamnose (Table 1). Based on the 16S rRNA gene sequence (1,383 bp) similarity, it was closely related to B. tequilensis 10bT with 99.64%, and, therefore, it was identified as B. tequilensis (Fig. 1) (Gatson et al., 2006).
 | Table 1. Differential phenotypic characteristics of the isolates. [Click here to view] |
Isolate KP1-10 showed irregular and cream colonies and grew in 0%–7% (w/v) NaCl, pH 6–8, and at 20°C–40°C. It showed positive for hydrolysis of arginine, gelatin and starch, citrate utilization, and nitrate reduction. Acid is produced from amygdalin, cellobiose, D-fructose, glycerol, D-maltose, mannose, melezitose, melibiose, rhamnose, D-ribose, salicin, sucrose, trehalose, and D-xylose but produced no acid for L-arabinose, galactose, inulin, lactose, mannitol, raffinose, and sorbitol (Table 1). Based on the 16S rRNA gene sequence (1,522 bp), it was closely related to B. cereus ATCC 14579T (Fig. 1) (98.91% sequence similarity) (Miller et al., 2016). Isolate KP1-14 had circular to slightly irregular and yellow colonies and grew in 0% (w/v)–10% (w/v) NaCl, pH 6–8, and at 20°C–40°C. It showed positive for hydrolysis of gelatin and arginine but negative for hydrolysis of starch, citrate utilization, and nitrate reduction. Acid is produced from amygdalin, cellobiose, D-fructose, galactose, glycerol, lactose, D-maltose, mannitol, mannose, melezitose, melibiose, raffinose, D-ribose, salicin, sucrose, and trehalose but negative for L-arabinose, inulin, rhamnose, sorbitol, and D-xylose. Based on the 16S rRNA gene sequence (1,376 bp) similarity, it was closely related to B. flexus NBRC 15715T with 99.93%. Therefore, it was identified as B. flexus (Priest et al., 1988) (Fig. 1).
 | Figure 1. Neighbor-joining tree based on the 16S rRNA gene sequences showing relationships among Bacillus and Staphylococcus isolates and related species. The numbers on the branches indicate the percentage bootstrap values of 1,000 replicates; only values >50% are indicated. Bar: 0.02 substitutions per nucleotide position. [Click here to view] |
 | Figure 2. Lipase activity (Unit/ml) of isolates in modified NB with 1% (v/v) Tween 80 or Tween 20 when p-NPB used as a substrate. [Click here to view] |
Isolate KP2-17 had circular and cream colonies and grew in 0%–5% (w/v) NaCl, pH 6–9, and at 20°C–40°C. It showed positive for hydrolysis of gelatin, starch, arginine, citrate utilization, nitrate reduction but negative for hydrolysis of Tween 80. Acid is produced from amygdalin, galactose, glycerol, lactose, D-maltose, D-ribose, sucrose, trehalose, and D-xylose but negative for L-arabinose, cellobiose, D-fructose, inulin, mannitol, mannose, melezitose, melibiose, raffinose, rhamnose, salicin, and sorbitol. Based on the 16S rRNA gene sequence similarity (1,351 bp), it was closely related to B. paramycoides NH24A2T with 99.48%; therefore, it was identified as B. paramycoides (Fig. 1) (Liu et al., 2017).
Staphylococcus isolates, KP3-03, KP4-02, KP4-08, KP5-02, KP5-03, and KP5-07 are Gram-reaction-positive cocci. Colonies were circular and white in color. They grew in pH 5.0–9.0, 0%–10% (w/v) NaCl, and at 25°C–40°C. They showed positive for citrate utilization and nitrate reduction but negative for hydrolysis of gelatin, starch, and arginine. The acid production was variable from lactose, D-ribose, trehalose, amygdalin, L-arabinose, cellobiose, D-fructose, galactose, glycerol, inulin, D-maltose, mannitol, mannose, melezitose, melibiose, raffinose, rhamnose, salicin, sorbitol, sucrose, and D-xylose. Based on the 16S rRNA gene sequence (1,390 bp), isolate KP4-02 was closely related to Staphylococcus cohnii subsp. urealyticus ATCC 49330T and was identified as Staphylococcus cohnii subsp. urealyticus with 100% sequence similarity to S. cohnii subsp. urealyticus (Kloos and Wolfshohl, 1991).
 | Figure 3. Lipase activity (Unit/ml) of isolates in modified NB with 1% (v/v) Tween 80 when p-NPP used as a substrate. [Click here to view] |
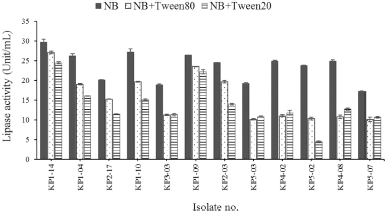
The isolates showed lipolytic activity when used p-NPB as a substrate in the culture medium when cultivated for 48 hours at 37°C in modified NB (17.27 ± 0.04 Unit/ml–29.68 ± 0.80 Unit/ml), modified NB supplemented with 1% (v/v) Tween 20 (4.41 ± 0.17 Unit/ml–24.5 ± 0.25 Unit/ml), and NB supplemented with 1% (v/v) Tween 80 (10.04 ± 0.56 Unit/ml–27.15 ± 0.33 Unit/ml) (Fig. 2). The highest lipase activity (29.68 ± 0.80 Unit/ml) was obtained from the culture medium of isolate KP1-14 (identified as B. flexus) cultured in modified NB medium.
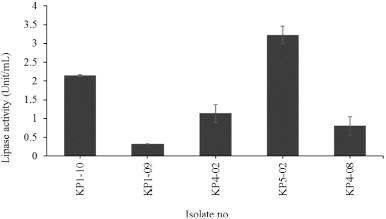
Moreover, the isolates KP1-10, KP1-09, KP4-02, KP5-02, and KP5-07 showed the lipase activity when used p-NPP as a substrate in the culture medium (modified NB supplemented with 1% (v/v) Tween 80) (0.32 ± 0.01 Unit/ml–3.22 ± 0.24 Unit/ml, Fig. 3). The highest lipase activity was found in S. cohnii subsp. urealyticus strain KP5-02. The high lipase activity is based on Tween 80 which contained the esters of oleic acid that are cleaved by lipases, whereas Tween 20 is easily hydrolyzed by esterases as it contained esters of lauric acid, a medium chain fatty acid (Kumar et al., 2012). The lipases display high activity toward the aggregated or insoluble substrates containing long chain fatty acids, whereas esterases activity is found to be highest toward the soluble substrate and broken ester bonds of short chain fatty acids (Fojan et al., 2000). After cultivated in modified NB supplemented with 1% (v/v) Tween 80, the result showed that the five isolates KP1-10, KP1-09, KP4-02, KP5-02, and KP5-07 hydrolyzed the substrate p-NPP (composed of long chain fatty acids, palmitic acid), which may be the result of their extracellular enzymes activity as true lipases that were induced by Tween 80. The hydrolysis of lipase is produced free fatty acids including low molecular weight volatile fatty acids such as acetic acid, propanoic acid, 2-methylpropanoic acid, butanoic acid, and 3-methylbutanoic acid (Sanceda et al., 2003). These free fatty acids are associated with the aroma and flavor in food products such as fish sauce (Nam-pla) and shrimp paste (Ka-pi) (Sharma et al., 2001). Therefore, these bacterial strains are interesting group of microorganisms that could be used as a source of enzyme in fermented foods for enhancement of aroma and flavor.
CONCLUSION
Twelve lipolytic bacterial strains from shrimp paste were identified as member of Bacillus and Staphylococcus based on their phenotypic characteristics and the 16S rRNA gene sequence analyses (98.91%–100% sequence similarity) and they were identified as B. tequilensis, B. infantis, B. flexus, B. paramycoides, Bacillus sp., and S. cohnii subsp. urealyticus. The strain B. flexus KP1-14 cultivated in NB medium and S. cohnii subsp. urealyticus strain KP5-02 cultivated in modified NB medium supplementary with 1% (v/v) Tween 80 showed the highest lipase activity by using p-nitrophenyl butyrate (C4) and p-nitrophenyl palmitate (C16), respectively. These strains will be useful for the application in fermentation of fish products in Thailand.
FINANCIAL SUPPORT
This study was supported by the Grant for International Research Integration: Research Pyramid, Ratchadaphiseksomphot Endowment Fund (GCURP_58_01_33_01), Chulalongkorn University as well as Chulalongkorn University. Additional thanks to the Ratchadapiseksomphot Endowment Fund, Chulalongkorn University for a post-doctoral fellowship to Dr. Rungsima Daroonpunt.
CONFLICT OF INTERESTS
The authors declare that there are no conflicts of interest.
REFERENCES
Abol-Fotouh DM, Bayoumi RA, Hassan MA. Production of thermoalkaliphilic lipase from Geobacillus thermoleovorans DA2 and application in leather industry. Enzyme Res, 2016; ID 9034364. CrossRef
Andualema B, Gessesse A. Microbial lipases and their industrial applications: review. Biotechnology, 2012; 11:100–18. CrossRef
Barrow GI, Feltham RKA. Cowan and Steel’s manual for the identification of medical bacteria. 3rd edition, Cambridge University Press, Cambridge, UK, 331 p, 1993. CrossRef
Camacho RM, Mateos JC, González-Reynoso O, Prado LA, Córdova J. Production and characterization of esterase and lipase from Haloarcula marismortui. J Ind Microbiol Biotechnol, 2009; 36:901–9. CrossRef
Chaijan M, Panpipat W. Darkening prevention of fermented shrimp paste by pre-soaking whole shrimp paste with pyrophosphate. As J Food Ag-Ind, 2012; 5(2):163–71.
Daroonpunt R, Tanasupawat S, Kudo T, Ohkuma M, Itoh T. Virgibacillus kapii sp. nov., isolated from Thai shrimp paste (Ka-pi). Int J Syst Evol Microbiol, 2016a; 66(4):1832–7. CrossRef
Daroonpunt R, Itoh T, Kudo T, Ohkuma M, Tanasupawat S. Bacillus piscicola sp. nov., isolated from Thai fish sauce (Nam-pla). Int J Syst Evol Microbiol, 2016b; 66(3):1151–5. CrossRef
Dey A, Chattopadhyay A, Saha P, Mukhopadhyay S. An approach to the identification and characterisation of a psychrotrophic lipase producing Pseudomonas sp. ADT3 from Arctic Region. Adv Biosci Biotechnol, 2014; 5:322–32. CrossRef
Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 1985; 39:783–91. CrossRef
Fojan P, Jonson PH, Petersen MTN, Petersen SB. What distinguishes an esterase from a lipase: a novel structural approach. Biochimie, 2000; 82(11):1033–41. CrossRef
Gatson JW, Benz BF, Chandrasekaran C, Satomi M, Venkateswaran K, Hart ME. Bacillus tequilensis sp. nov., isolated from a 2000-year-old Mexican shaft-tomb, is closely related to Bacillus subtilis. Int J Syst Evol Microbiol, 2006; 56:1475–84. CrossRef
Garcia-Lepe R, Neuro OM, Reyes F, Sanatamaría F. Lipases in autolysed cultures of filamentous fungi. Lett Appl Microbiol, 1997; 25:127–30. CrossRef
Gayathri VR, Perumal P, Mathew LP, Prakash B. Screening and molecular characterization of extracellular lipase producing Bacillus species from coconut oil mill soil. Int J Sci Technol, 2013; 2:502–9.
Ghasemi Y, Rasoul-Amini S, Ebrahiminezhad A, Zarrini G, Kazemi A, Mousavi-Khorshidi S, Ghoshoon MB, Raee MJ. Halotolerant amylase production by a novel bacterial strain, Rheinheimera aquimaris. Res J Microbiol, 2010; 5(2):144–9. CrossRef
Gordon RE. The genus Bacillus. In: Laskin AI, Lechevalier HA (eds.). Hand-book of Microbiology, Organismic Microbiology. The Chemical Rubber Co. Press, Cleveland, vol. 1, pp. 71–88, 1973.
Itou K, Kobayashi S, Ooizumi T, Akahane Y. Changes of proximate composition and extractive components in narezushi, a fermented mackerel product, during processing. Fish Sci, 2006; 72:1269–76. CrossRef
Javed S, Azeem F, Hussain S, Rasul I, Siddique MH, Riaz M, Afzal M, Kouser A, Nadeem H. Bacterial lipases: a review on purification and characterization. Prog Biophys Mol Biol, 2018; 132:23–34. CrossRef
Kloos WE, Wolfshohl JF. Staphylococcus cohnii subspecies: Staphylococcus cohnii subsp. cohnii subsp. nov. and Staphylococcus cohnii subsp. urealyticum subsp. nov. Int J Syst Bacteriol, 1991; 41(2):284–9. CrossRef
Ko KS, Oh WS, Lee MY, Lee JH, Lee H, Peck KR, Lee NY, Song JH. Bacillus infantis sp. nov. and Bacillus idriensis sp. nov., isolated from a patient with neonatal sepsis. Int J Syst Evol Microbiol, 2006; 56:2541–4. CrossRef
Kumar R, Sharma A, Kumar A, Singh D. Lipase from Bacillus pumilus RK31: production, purification and some properties. World Appl Sci J, 2012; 16(7):940–8.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 2016; 33(7):1870–4. CrossRef
Lee DW, Koh SY, Kim KJ, Kim BC, Choi HJ, Kim DS, Suhartono MT, Pyun YR. Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiol Lett, 1999; 179(2):393–400. CrossRef
Leifson E. Determination of carbohydrate metabolism of marine bacteria. J Bacteriol, 1963; 85:1183–4.
Li X, Qian P, Wu SG, Yu HY. Characterization of an organic solvent-tolerant lipase from Idiomarina sp. W33 and its application for biodiesel production using Jatropha oil. Extremophiles, 2014; 18:171–8. CrossRef
Liu Y, Chen D, Yan Y, Peng C, Xu L. Biodiesel synthesis and conformation of lipase from Burkholderia cepacia in room temperature ionic liquids and organic solvents. Bioresour Technol, 2011; 102:10414–8. CrossRef
Liu Y, Lai Q, Zeng R, Ye D, Xu J, Shao Z. Proposal of nine novel species of the Bacillus cereus group. Int J Syst Evol Microbiol, 2017; 67(8):2499–508. CrossRef
Miller RA, Beno SM, Kent J, Carroll LM, Martin NH, Boor KJ, Kovac J. Bacillus wiedmannii sp. nov., a psychrotolerant and cytotoxic Bacillus cereus group species isolated from dairy foods and dairy environments. Int J Syst Evol Microbiol, 2016; 66:4744–53. CrossRef
Phoottosavako M, Keeratipibul S, Techo S, Tanasupawat S. Identification and characterization of lipolytic bacteria from Thai fermented foods. Mal J Microbiol, 2015; 11(3):231–9. CrossRef
Pongsetkul J, Benjakul S, Sumpavapol P, Vongkamjan K, Kazufumi O. Bacillus subtilis K-C3 isolated from Thai salted shrimp paste (Kapi): Its extracellular enzymes and use as a starter culture in Kapi production. J Food Biochem, 2018; 42(6); doi:10.1111/jfbc.12649. CrossRef
Priest FG, Goodfellow M, Todd C. A numerical classification of the genus Bacillus. J Gen Microbiol, 1988; 134:1847–82. CrossRef
Priji P, Unni KN, Sajith S, Binod P, Benjamin S. Production, optimization, and partial purification of lipase from Pseudomonas sp. strain BUP6, a novel rumen bacterium characterized from Malabari goat. Biotechnol Appl Biochem, 2015; 62(1):71–8. CrossRef
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 1987; 4:406–25.
Sanceda NG, Suzuki E, Kurata T. Branched chain amino acids as source of specific branched chain volatile fatty acids during the fermentation process of fish sauce. Amino Acids, 2003; 24:81–7.
Sharma R, Chisti Y, Banerjee UC. Production, purification, characterization, and applications of lipases. Biotechnol Adv, 2001; 19(8):627–62. CrossRef
Wittanalai S, Rakariyatham N, Deming RL. Volatile compounds of vegetarian soybean kapi, a fermented Thai food condiment. Afr J Biotechnol, 2011; 10:821–30.
Yamada Y, Katsura K, Kawasaki K, Widyastuti Y, Saono S, Seki T, Uchimura T, Komagata K. Asaia bogorensis gen. nov., sp. nov., an unusual acetic acid bacterium in the α-Proteobacteria. Int J Syst Evol Microbiol, 2000; 50:823–9. CrossRef
Yang W, He Y, Xu L, Zhang H, Yan Y. A new extracellular thermo-solvent-stable lipase from Burkholderia ubonensis SL-4: Identification, characterization and application for biodiesel production. J Mol Catal B Enzym, 2016; 126:76–89. CrossRef
Yele VU, Desai KA. New thermostable and organic solvent-tolerant lipase from Staphylococcus warneri; optimization of media and production conditions using statistical methods. Appl Biochem Biotechnol, 2015; 175:855–69. CrossRef
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol, 2017; 67:1613–7. CrossRef